Abstract
Genome-wide association studies have identified risk loci linked to inflammatory bowel disease (IBD)1—a complex chronic inflammatory disorder of the gastrointestinal tract. The increasing prevalence of IBD in industrialized countries and the augmented disease risk observed in migrants who move into areas of higher disease prevalence suggest that environmental factors are also important determinants of IBD susceptibility and severity2. However, the identification of environmental factors relevant to IBD and the mechanisms by which they influence disease has been hampered by the lack of platforms for their systematic investigation. Here we describe an integrated systems approach, combining publicly available databases, zebrafish chemical screens, machine learning and mouse preclinical models to identify environmental factors that control intestinal inflammation. This approach established that the herbicide propyzamide increases inflammation in the small and large intestine. Moreover, we show that an AHR–NF-κB–C/EBPβ signalling axis operates in T cells and dendritic cells to promote intestinal inflammation, and is targeted by propyzamide. In conclusion, we developed a pipeline for the identification of environmental factors and mechanisms of pathogenesis in IBD and, potentially, other inflammatory diseases.
Zebrafish offer multiple advantages for immune-related small-molecule screens because they reproduce in high numbers, are transparent during early developmental stages and have an immune system that shares important features with its human counterpart3–6. 2,4,6-trinitrobenzenesulfonic acid (TNBS) induces experimental T cell-driven colitis in mice7, and is also reported to induce intestinal inflammation in zebrafish8. Indeed, treatment of zebrafish larvae at 7 days post-fertilization (d.p.f.) with 25 μg ml−1 TNBS resulted in the loss of crypt and villi morphology, expansion of the gut lumen and decreased gut peristaltic movement (Fig. 1a and Extended Data Fig. 1a,b), concomitant with increased expression of the pro-inflammatory genes il17a/f, tnfa, ifng, il1b and nos2a (Fig. 1b). Pharmacological inhibition of nitric oxide synthase (NOS) or activation of the aryl hydrocarbon receptor (AHR) with 6-formylindolo[3,2-b]carbazole (FICZ) ameliorated intestinal inflammation in zebrafish (Fig. 1a–c and Extended Data Fig. 1c), recapitulating previous observations in mouse TNBS-induced colitis9. Moreover, treatment with lenaldekar, an inhibitor of T cell activation in zebrafish, mice and humans, or the use of lymphocyte-deficient rag1- or rag2-mutant zebrafish decreased TNBS-induced intestinal inflammation (Extended Data Fig. 1d–k), suggesting that T cells contribute to intestinal pathology induced by TNBS in 7 d.p.f. zebrafish.
Fig. 1 |. Identification of environmental chemicals that boost intestinal inflammation.
a, Zebrafish (7 d.p.f.) were immersed in TNBS-containing E3 medium to induce intestinal inflammation and were scored 72 h later. The images show the effect of NOS inhibition (NOSi) and AHR activation by FICZ on intestinal pathology. The plate, microscope and slide elements of this figure were created using BioRender. b, Gene expression in naive or TNBS-treated zebrafish exposed to vehicle, NOSi (5 μM) or FICZ (5 μM). n = 12 (il17a/f and ifng) and n = 14 (tnfa, il1b and nos2a). c, Intestinal scores of naive or TNBS-treated zebrafish exposed to vehicle, NOSi or FICZ (top), or environmental chemicals (0.2, 1, 5 or 20 μM) that suppressed (middle) or promoted (bottom) inflammation. n = 24 per group. d, Gene expression in TNBS-treated zebrafish exposed to candidate chemicals (1 or 20 μM). n = 9 per group. e, The workflow for the identification of candidate environmental chemicals based on the US EPA ToxCast database. f, The top 20 candidate chemicals predicted to worsen intestinal inflammation identified by the RF–RWR algorithm. g, Intestinal scores in TNBS-treated zebrafish exposed to environmental chemicals (0.2, 1, 5 or 20 μM) identified by the RF–RWR algorithm. n = 12 per group. Grey shading indicates lethality. h,i, Intestinal score (h; n = 48 per group) and gene expression (i; n = 18 per group) in vehicle- or propyzamide-treated (0.2, 1, 5 or 20 μM) TNBS zebrafish. For b, h and i, data are mean ± s.e.m. Statistical analysis was performed using one-way analysis of variance (ANOVA) followed by Dunnett’s multiple-comparisons test (b–d and g–i); ****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05.
To screen for environmental chemicals that modulate intestinal inflammation, we established a platform that integrates IBD genetics, high-throughput toxicology and small-molecule screens in the TNBS-induced colitis zebrafish model. In these studies, we used the EPA ToxCast database, which collects data on the activity of chemicals used in industry, agriculture and consumer products in high-throughput biochemical and cell-based assays10.
We first examined the ToxCast database to identify environmental chemicals that are active in bioassays associated with tumour necrosis factor (TNF), interferon (IFN), interleukin-1β (IL-1β), JAK/STAT, peroxisome proliferator-activated receptor (PPAR) and AHR signalling, all of which have been linked to intestinal inflammation. We identified 111 chemicals in the ToxCast database that met these criteria (Extended Data Fig. 1l and Supplementary Tables 1 and 2), and these were selected for further evaluation in the zebrafish model of TNBS-induced intestinal inflammation. Of these candidate chemicals, 62 were excluded due to lethality or the induction of overt morphological changes. The remaining 49 chemicals were screened at 20 μM, as previously done to test environmental chemicals5,6,11–13 (Supplementary Table 3). Chemicals active in the zebrafish model of TNBS-induced intestinal inflammation were retested at 20 μM, 5 μM, 1 μM and 0.2 μM, resulting in the identification of 4 chemicals that suppressed and 13 chemicals that boosted zebrafish intestinal pathology (Fig. 1c,d).
Candidate chemicals boosting intestinal inflammation
Machine learning methods have been used to build predictive models of the biological activity of chemicals14. Thus, to identify additional environmental chemicals in the ToxCast database that modulate intestinal inflammation, we used a machine learning approach based on a training set that included the 13 chemicals that boosted intestinal inflammation, the 4 chemicals that ameliorated it and 13 representative chemicals that had no effect in our initial zebrafish screen (Fig. 1c–e and Supplementary Table 3). We first eliminated chemicals and bioassays that were poorly correlated with the set of experimentally validated chemicals, or that contained missing values; this resulted in a testing set of 297 chemicals and 233 bioassays (Fig. 1e). We then used a Kruskal–Wallis test to identify the ToxCast bioassays that best discriminated IBD-promoting, IBD-ameliorating and no-effect chemicals in the training set, identifying 16 bioassays that we defined as an IBD bioactivity feature (Fig. 1e, Extended Data Fig. 1m and Supplementary Tables 4 and 5). On the basis of this IBD bioactivity feature, we built a random forest (RF) model to identify in the ToxCast database additional chemicals predicted to worsen intestinal inflammation, using a random walk with restart (RWR) algorithm to rank them (Fig. 1f and Supplementary Table 6).
To validate the RF–RWR results, we evaluated the top 20 chemicals predicted to be pro-inflammatory (Fig. 1f,g); 6 out of 8 chemicals (75%) that were non-lethal at 20 μM boosted TNBS-induced intestinal inflammation in zebrafish. By contrast, when we evaluated 40 chemicals randomly selected from the ToxCast database, only 4 out of the 22 chemicals that were non-lethal worsened inflammation (18%), suggesting that the machine learning algorithm significantly enriched for chemicals that boost intestinal inflammation (P = 0.0072, χ2 test; Extended Data Fig. 1n). Similarly, 6 out of 13 (46%) and 6 out of 20 (30%) predicted IBD-worsening chemicals that were non-lethal at 5 μM and 1 μM, respectively, boosted intestinal inflammation, compared with only 5 out of 30 (16%) and 2 out of 37 (5%) of random compounds tested at 5 μM and 1 μM, respectively (P = 0.0209 (5 μM) and P = 0.0054 (1 μM), χ2 test; Fig. 1g and Extended data Fig. 1n); no significant enrichment was detected at 0.2 μM.
The top 20 candidates identified using the RF–RWR approach included 11 agriculture-related chemicals (Fig. 1f and Supplementary Table 6). We selected propyzamide for follow-up studies because it is used on fruits, vegetables and other crops, and also in ornamental gardens. Propyzamide boosted TNBS-induced intestinal inflammation in zebrafish, as indicated by the analysis of intestinal scores and the expression of pro-inflammatory genes (Fig. 1g–i). However, propyzamide administration did not induce intestinal inflammation in the absence of TNBS (Extended Data Fig. 1o,p), indicating that propyzamide has a role in the amplification but not the initiation of gut pathology. Interestingly, AHR activation by FICZ or NOS inhibition partially interfered with the worsening of TNBS-induced colitis by propyzamide (Extended Data Fig. 1q).
Propyzamide identifies regulators of gut inflammation
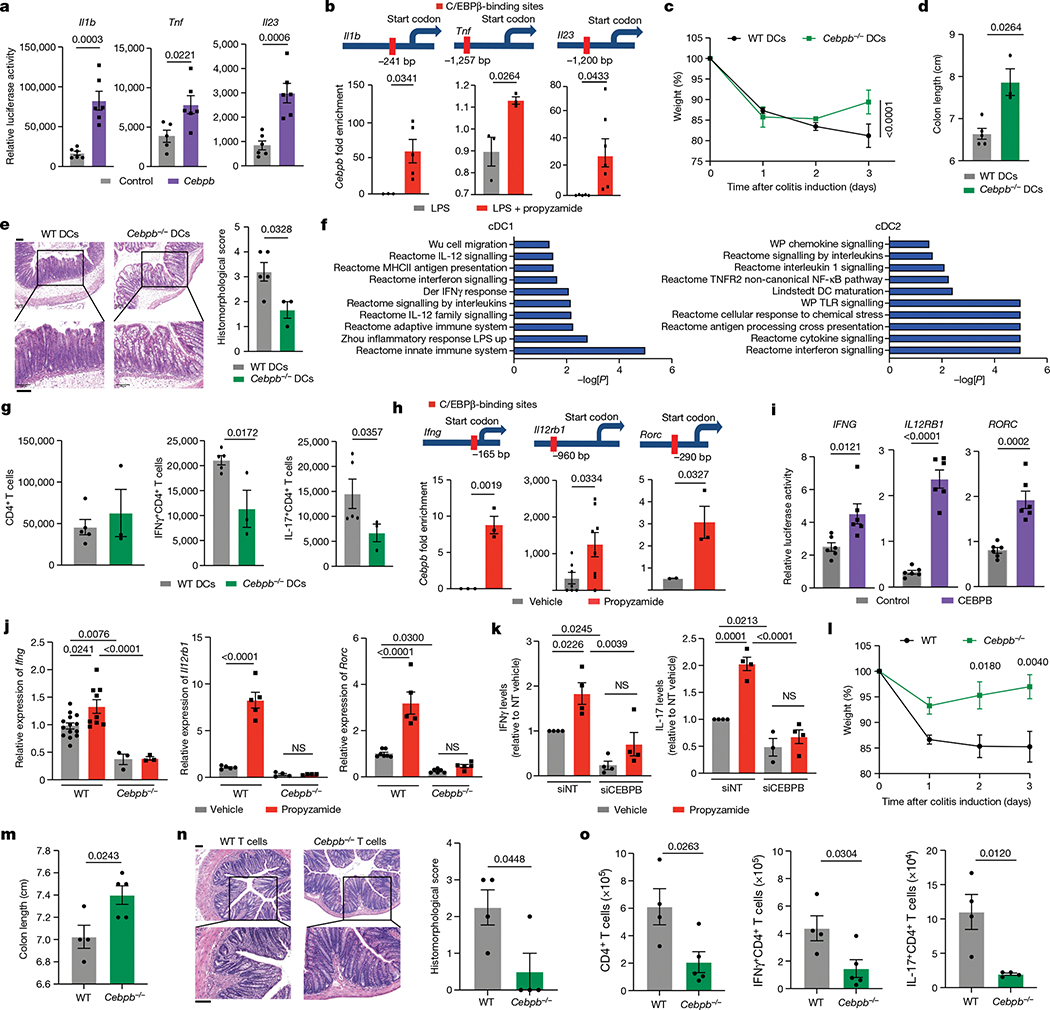
Environmental chemicals that modulate inflammation can be used as probes to search for mechanisms of disease pathogenesis and candidate therapeutic targets, as we recently described in the context of neuroinflammation5. Thus, to identify immunoregulatory mechanisms targeted by propyzamide in mammals we first used the mouse model of TNBS-induced colitis. Propyzamide worsened intestinal pathology as indicated by weight loss, colon shortening and histological evaluation (Fig. 2a–c). Propyzamide also increased IL-17+CD4+ T cells, IL-17+RORγt+CD4+ T cells, IFNγ+CD4+ T cells and IFNγ+CD8+ T cells in the colon (Fig. 2d and Extended Data Fig. 2a–e), but did not modify IL-17 production by CD8+ T cells and γδ T cells (Extended Data Fig. 2f). Propyzamide administration in the absence of TNBS did not alter weight, colon length, CD4+ T cells, CD8+ T cells, γδ T cells or innate lymphoid cell (ILC) numbers (Extended Data Fig. 2g–m), in agreement with a role for propyzamide in the amplification, but not the initiation, of intestinal inflammation.
Fig. 2 |. Propyzamide identifies candidate regulators of intestinal inflammation.
a,b, Weight (a) and colon length (b) of naive or vehicle- or propyzamide-treated (100 mg kg−1) TNBS mice. n = 5 (naive), n = 6 (TNBS + vehicle) and n = 10 (TNBS + propyzamide). Statistical analysis was performed using two-way ANOVA with Tukey’s multiple-comparison post hoc test. c, Haematoxylin and eosin (H&E) colon images and histomorphology scores. n = 5 mice per group. The arrows indicate leukocyte infiltration. Scale bars, 100 μm. d, Colonic IFNγ+CD4+, IL-17+CD4+ and IFNγ+CD8+ T cells in vehicle- or propyzamide-treated TNBS mice. n = 6 (vehicle) and n = 5 (propyzamide). e,f, Heat map (e) and ingenuity pathway analysis (IPA) (f) showing mRNA expression determined by bulk RNA-seq in colons from vehicle-treated (n = 2) or propyzamide-treated (n = 3) TNBS mice. Sig, signalling. g, AHR inhibition of NF-κB–C/EBPβ signalling as a candidate mediator of propyzamide effects in TNBS mice. h, Rela and Cebpb expression determined by qPCR in colonic CD45+ cells from vehicle-treated (n = 4) or propyzamide-treated (n = 5) TNBS mice. Statistical analysis was performed using two-tailed Mann–Whitney U-tests. i, Uniform manifold approximation and projection (UMAP) plot of a scRNA-seq analysis of colon cells from propyzamide- or vehicle-treated naive and TNBS mice. n = 5 mice per group. j, DCs and T cells from vehicle- or propyzamide-treated mice within each subpopulation. k, Pathways upregulated by propyzamide treatment. l, UMAP plot of the DC cluster in Fig. 3i. m, Cells from vehicle- or propyzamide-treated mice within each subpopulation. n, AHR signature score M9986 in DCs. Statistical analysis was performed using unpaired two-tailed t-tests. o, Jejunal weight/length ratio before and 3 h and 24 h after administration of anti-CD3 antibodies. n = 10 (vehicle, naive), n = 15 (vehicle, 3 h after anti-CD3 administration), n = 6 (vehicle, 24 h after anti-CD3), n = 9 (propyzamide, naive), n = 14 (propyzamide, 3 h after anti-CD3) and n = 12 (propyzamide, 24 h after anti-CD3 injection) mice. p, H&E staining of jejunum sections 24 h after injection of anti-CD3 antibodies. The villus height and villus height/crypt length ratio are shown. Scale bars, 100 μm. q, TUNEL staining and TUNEL+ cells in the jejunum 24 h after anti-CD3 administration. Scale bar, 100 μm. For a, b, d, h and o–q, data are mean ± s.e.m.
Dysbiosis contributes to IBD pathology, and human-made chemicals can perturb the microbiome, affecting intestinal inflammation and metabolism15. After oral administration, propyzamide was absorbed, detected in the plasma and quickly metabolized; the induction of colitis with TNBS did not affect propyzamide absorption or elimination (Extended Data Fig. 2n,o). We did not detect propyzamide in bile acids, but low levels of non-metabolized propyzamide were detected in the faeces, suggesting that it interacts with the intestinal flora (Extended Data Fig. 2n,o). Indeed, propyzamide administration reduced microbiome diversity in the ileum and the caecum, and altered microbiome composition as indicated by the analysis of the Pielou evenness index of α-diversity and β-diversity in 16S rRNA sequences (Extended Data Fig. 3a,b and Supplementary Tables 7 and 8). Moreover, in propyzamide-treated TNBS mice, we detected the expansion of Sutterellaceae (Extended Data Fig. 3c,d), which is linked to intestinal inflammation16. Thus, we studied the contribution of propyzamide-induced dysbiosis to intestinal inflammation in faecal-microbiota-transfer studies17. Germ-free mouse recipients of faecal microbiota from propyzamide- or vehicle-treated mice developed comparable intestinal inflammation after TNBS administration (Extended Data Fig. 3e–j), indicating that, although propyzamide alters the intestinal microbiota, intestinal dysbiosis does not contribute to its effects on intestinal inflammation.
Using RNA-sequencing (RNA-seq) analyses, we detected the upregulation of pro-inflammatory pathways, including those associated with leukocyte extravasation, integrin signalling and NF-κB activation, in colons from propyzamide-treated TNBS mice (Fig. 2e,f). An analysis of upstream regulators in the RNA-seq dataset identified decreased AHR suppression of NF-κB-driven C/EBPβ as a candidate mechanism for mediating propyzamide-induced transcriptional effects (Fig. 2g). Indeed, quantitative PCR (qPCR) validation studies detected increased expression of Rela (encoding the p65 subunit of NF-κB) and Cebpb (Fig. 2h), and the upregulation of pro-inflammatory Tnf, Il1b, Il23 and Il6 in lamina propria mononuclear cells (LPMCs); no changes were detected in Il10 or Tgfb expression (Extended Data Fig. 4a,b).
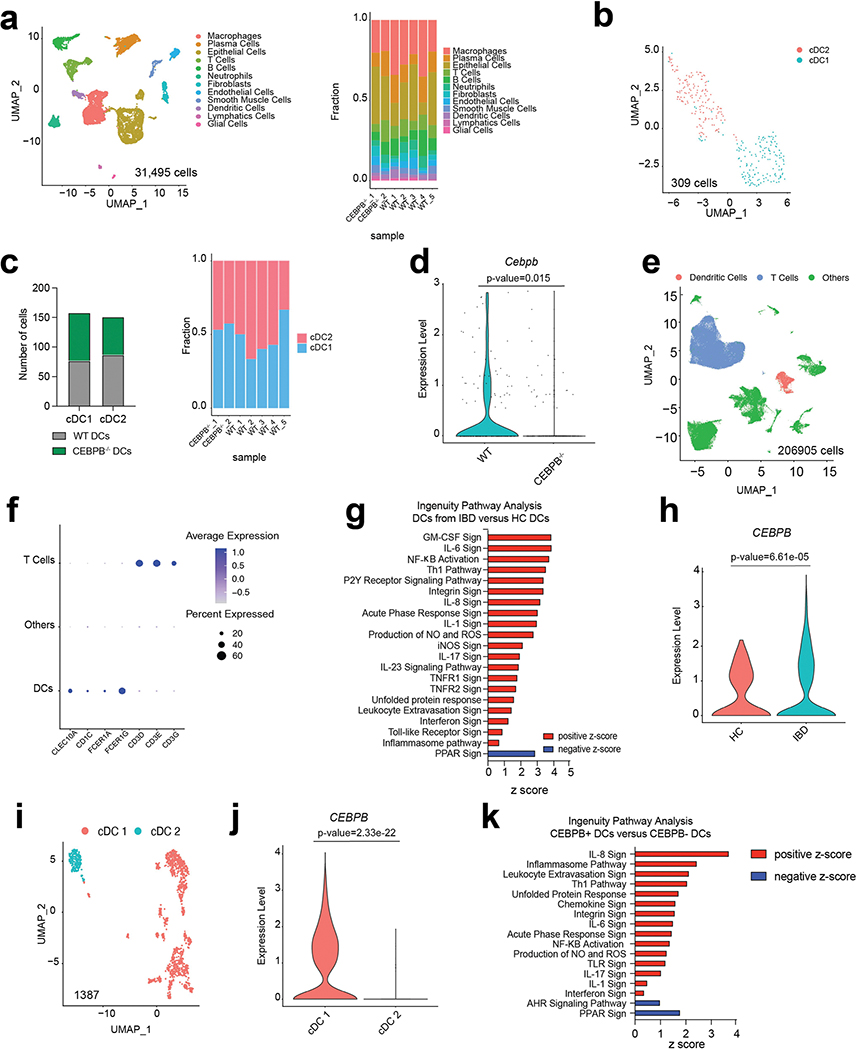
To further investigate the mechanism by which propyzamide worsens intestinal inflammation, we analysed the colons of propyzamide-treated mice using single-cell RNA-seq (scRNA-seq) (Fig. 2i and Extended Data Fig. 4c,d). Propyzamide administration to TNBS mice resulted in the expansion of the T cell and dendritic cell (DC) clusters, concomitant with the upregulation of NF-κB-driven C/EBPβ pro-inflammatory gene expression and decreased AHR signalling (Fig. 2j–n and Extended Data Fig. 4d–h). These effects of propyzamide on T cells and DCs were not detected after propyzamide administration in the absence of TNBS (Extended Data Fig. 4d–i).
Finally, we investigated whether propyzamide also boosts inflammation in the small intestine using the model of T cell-driven enteritis induced by anti-CD3 monoclonal antibodies7. Propyzamide administration worsened intestinal pathology and T cell-driven inflammation (Fig. 2o–q). Moreover, in agreement with our findings in TNBS-induced colitis, transcriptomic analyses detected decreased AHR signalling concomitant with increased NF-κB activation and expression of Cebpb and other pro-inflammatory molecules, as well as increased IL-17 and IFNγ production by CD4+ T cells (Extended Data Fig. 4j–o). Propyzamide administration in the absence of anti-CD3 did not affect the number of small intestine immune cells or their transcriptional programs (Extended Data Fig. 5a–e). Together, these findings suggest that propyzamide worsens inflammation by acting on the AHR–NF-κB–C/EBPβ signalling axis that regulates intestinal immune cell responses in both the large and small intestine.
Propyzamide inhibits AHR signalling
The transcription factor AHR modulates intestinal homeostasis and inflammation18, and has been shown to suppress NF-κB activation19. On the basis of the decreased AHR signalling and increased NF-κB activation detected in DCs and T cells from propyzamide-treated mice (Fig. 2j–n and Extended Data Fig. 4j–o), we studied the effects of propyzamide on AHR. In reporter assays, propyzamide reduced FICZ-induced AHR activation (Fig. 3a), but did not interfere with the activation of the nuclear receptors RARα or PPARα (Extended Data Fig. 5f,g). Moreover, propyzamide suppressed FICZ-induced expression of the AHR-target genes Cyp1a1 and Cyp1b1 in human and mouse primary DCs and T cells (Fig. 3b–e). Indeed, propyzamide reduced the binding of the ligand 3H-TCDD to mouse and human AHR in cell-free binding assays (Fig. 3f).
Fig. 3 |. Propyzamide suppresses AHR signalling.
a, AHR reporter activity after propyzamide (10 μM) and/or FICZ (0.5 μM) treatment. n = 5 (FICZ and propyzamide + FICZ), n = 6 (other groups). b, Cyp1a1 and Cyp1b1 expression in wild-type (WT) or AHRd DCs. n = 6 (vehicle), n = 10 (propyzamide), n = 5 (FICZ), n = 8 (propyzamide + FICZ) for Cyp1a1 in wild-type DCs; n = 3 (propyzamide and propyzamide + FICZ) and n = 5 (FICZ) for Cyp1b1 in wild-type DCs; n = 3 (vehicle and propyzamide) for Cyp1b1 in AHRd DCs; and n = 4 (other groups). c, CYP1A1 expression in human DCs. n = 6 donors. d, Cyp1a1 and Cyp1b1 expression in mouse T cells. n = 4 (propyzamide Cyp1b1; vehicle, propyzamide and FICZ Cyp1a1), n = 3 (other groups). e, CYP1A1 expression in human T cells. n = 4 donors. f, 3H-TCDD-displacement assay from human or mouse AHR. n = 3 independent experiments. g–j, Colon length (g), histopathology score (h), representative images of colon (i) and the numbers of IFNγ+CD4+ and IL-17+CD4+ T cells ( j) from wild-type (n = 5 (vehicle) and n = 6 (propyzamide)) and AHRd TNBS-colitis mice (n = 3 per group). Scale bars, 100 μm. k–m, NF-κB p65 phosphorylation (k) (n = 4 wild-type, n = 3 AHRd) and Cebpb expression in CD4+ T cells (l) and DCs (m) from wild-type and AHRd mice during TNBS-induced colitis (n = 10 and n = 8 (wild-type vehicle T cells and DCs, respectively), n = 4 and n = 8 (wild-type propyzamide T cells and DCs, respectively) and n = 3 (other groups). n, Cebpb expression in DCs treated with vehicle (n = 13), propyzamide (n = 6), FICZ (n = 10) or propyzamide + FICZ (n = 14). o, Il1b, Il23 and Tnf expression in wild-type or Cebpb−/− DCs (n = 11 (Il1b and Tnf, vehicle, wild-type DCs), n = 6 (Il1b, propyzamide, wild-type DCs), n = 4 (Cebpb−/− DCs, vehicle), n = 8 (Il23, propyzamide, wild-type DCs) and n = 5 (other groups). p, IL1B, IL23 and TNF expression in human DCs treated with propyzamide after CEBPB (siCEBPB) or control (siNT) knockdown. n = 4 donors per condition. Statistical analysis was performed using one-way ANOVA with Dunnett’s, Sidak’s or Tukey’s post hoc test for selected multiple comparisons (a, b, d, f–h and k–p) and unpaired two-tailed t-tests (c and e). For a–h and j–p, data are mean ± s.e.m.
To evaluate the effects of propyzamide on AHR signalling in vivo we used mice carrying the Ahr d allele (AHRd mice), which contains mutations that decrease its interactions with physiological ligands20. In agreement with the reported protective effects of AHR in TNBS-induced colitis9, AHRd mice displayed worsened TNBS-induced colitis, increased IFNγ+CD4+ and IL-17+CD4+ T cells in the colon, and upregulation of pro-inflammatory molecule expression in CD4+ T cells and DCs, which was not worsened by propyzamide (Fig. 3g–j and Extended Data Fig. 5h,i). Moreover, AHRd CD4+ T cells and DCs displayed increased phosphorylation of the NF-κB p65 subunit, and this increase in p65 phosphorylation was not boosted by propyzamide (Fig. 3k). Collectively, these findings suggest that propyzamide interferes with the suppression of NF-κB activation by AHR.
C/EBPβ boosts pro-inflammatory DC function
Our transcriptional analyses identified NF-κB-driven C/EBPβ signalling as a candidate mediator of the pro-inflammatory effects of propyzamide (Fig. 2g). Propyzamide upregulated Cebpb expression in DCs in an NF-κB-dependent manner (Fig. 3l,n and Extended Data Figs. 5j–l and 10a,c); AHR activation with FICZ decreased Cebpb expression, whereas DCs isolated from AHRd mice showed higher Cebpb expression. Notably, CD4+ T cells isolated from AHRd mice also showed higher Cebpb expression (Fig. 3m). Interestingly, propyzamide boosted NF-κB activation in primary splenic DCs from p65 reporter mice but did not induce mouse microtubule destabilization (Extended Data Fig. 5l,m), a process triggered by propyzamide in plants21 which has been linked to NF-κB activation22.
Propyzamide boosted, in a C/EBPβ-dependent manner, Il1b, Tnf and Il23 expression in LPS-activated mouse splenic and bone-marrow-derived dendritic cells (BMDCs), and also in human DCs (Fig. 3o,p and Extended Data Fig. 6a–e). Indeed, in publicly available chromatin immunoprecipitation followed by high-throughput sequencing (ChIP–seq) datasets23, we detected C/EBPβ binding to the promoters of Il1b, Il23 and Tnf (Extended Data Fig. 6f); these promoters were activated by propyzamide in reporter assays in DCs (Fig. 4a,b).
Fig. 4 |. NF-κB–C/EBPβ signalling boosts colitogenic T cell responses.
a, Transactivation of Il1b, Il23 and Tnf promoters after Cebpb expression in DC2.4 cells. n = 6 per condition. b, Predicted C/EBPβ-binding sites (top) and C/EBPβ recruitment (bottom) to Il1b, Il23 and Tnf promoters in DCs. n = 3–7 per group. c,d, Weight change (c) and colon length (d) in wild-type and Cebpb−/− DC chimeras. e, Representative H&E colon images and histomorphology scores. n = 5 (wild-type DCs) and n = 3 (Cebpb−/− DCs). Scale bar, 100 μm. f, GSEA showing pathways downregulated in cDC1 and cDC2 from Cebpb−/− DC chimeras. g, Total CD4+, IFNγ+CD4+ and IL-17+CD4+ T cells in wild-type DC (n = 5 mice) and Cebpb−/− DC (n = 3 mice) chimeras. Statistical analysis was performed using unpaired t-tests. h, Predicted C/EBPβ-binding sites (top) and C/EBPβ recruitment (bottom) to Ifng, Il12rb1 and Rorc promoters in mouse CD4+ T cells. n = 3–8 per group. i, Transactivation of IFNG, IL12RB1 and RORC promoters by C/EBPβ expression in human CD4+ T cells. n = 6 per condition. j, Ifng, Il17, Rorc and Il12rb1 expression in splenic wild-type or Cebpb−/− CD4+ cells treated with propyzamide in vitro. Ifng: n = 14 (wild-type, vehicle), n = 9 (wild-type, propyzamide) and n = 3 (Cebpb−/−); Rorc: n = 7 (wild-type, vehicle), n = 6 (Cebpb−/−, vehicle) and n = 5 (other groups); Il12rb1: n = 5 (wild-type, vehicle and propyzamide) and n = 4 (other groups). NS, not significant. k, IFNγ and IL-17 levels in human T cell supernatants after CEBPB silencing and propyzamide treatment. n = 3 (Il17, siCEBPB, vehicle) and n = 4 (other groups). l,m, Weight change (l) and colon length (m) in Rag1-KO mice reconstituted with wild-type or Cebpb−/− T cells during TNBS-induced colitis. n, Representative H&E colon staining and histomorphology scores. n = 4 (wild-type) and n = 5 (Cebpb−/−). Scale bars, 100 μm. o, Total CD4+, IFNγ+CD4+ and IL-17+CD4+ T cells from colons of the TNBS-treated mice in l. n = 5 (CD4+ and IFNγ+CD4+, Cebpb−/−) and n = 4 (other groups) mice. Statistical analysis was performed using one-way ANOVA with Dunnett’s, Sidak’s, Holm Sidak’s or Tukey’s post hoc test for selected multiple comparisons (c, d and j–m). For a–e and g–o, data are mean ± s.e.m.
To evaluate the functional effects of NF-κB–C/EBPβ signalling, we analysed ex vivo DCs isolated from propyzamide- and vehicle-treated mice. Splenic DCs from propyzamide-treated mice showed an increased ability to activate OVA323–339-specific OT-II CD4+ T cells and promote T helper 1 (TH1) and TH17 cell polarization (Extended Data Fig. 6g,h). Similar results were obtained when we treated in vitro primary mouse and human DCs with propyzamide (Extended Data Fig. 6i,j). Interestingly, C/EBPβ inactivation in Cebpb-knockout mice or after short interfering RNA (siRNA) knockdown of Cebpb reduced effector T cell activation and polarization in both propyzamide- and vehicle-treated DCs, suggesting that NF-κB–C/EBPβ participates in the physiological regulation of DC function (Extended Data Fig. 6i,j). Indeed, C/EBPβ overexpression using a cumate-inducible system upregulated Il1b, Tnf and Il23 expression in primary mouse splenic DCs (Extended Data Fig. 6k,l).
To investigate the role of C/EBPβ expressed in DCs during intestinal inflammation, we generated mouse chimeras with C/EBPβ-deficient classical DCs (cDCs)24. In brief, we reconstituted lethally irradiated wild-type mice with bone marrow cells expressing the diphtheria toxin receptor under the control of the Zbtb46 promoter25. In these mice, DTR+ cDCs can be depleted by chronic administration of diphtheria toxin without the lethality observed in Zbtb46-DTR mice25, and then replenished with wild-type or C/EBPβ-deficient DC precursors to generate wild-type DC or Cebpb−/− DC mice, respectively. C/EBPβ deficiency in cDCs resulted in the amelioration of TNBS-induced colitis (Fig. 4c–e). We detected similar cDC1 and cDC2 numbers in wild-type DC and Cebpb−/− DC mice but, in scRNA-seq analyses, C/EBPβ-deficient DCs displayed decreased activation of pro-inflammatory pathways, such as those linked to response to interferon and interleukin signalling (Fig. 4f and Extended Data Fig. 7a–d). We also detected a decrease in TH1 and TH17 cell numbers in the colons of Cebpb−/− DC mice (Fig. 4g).
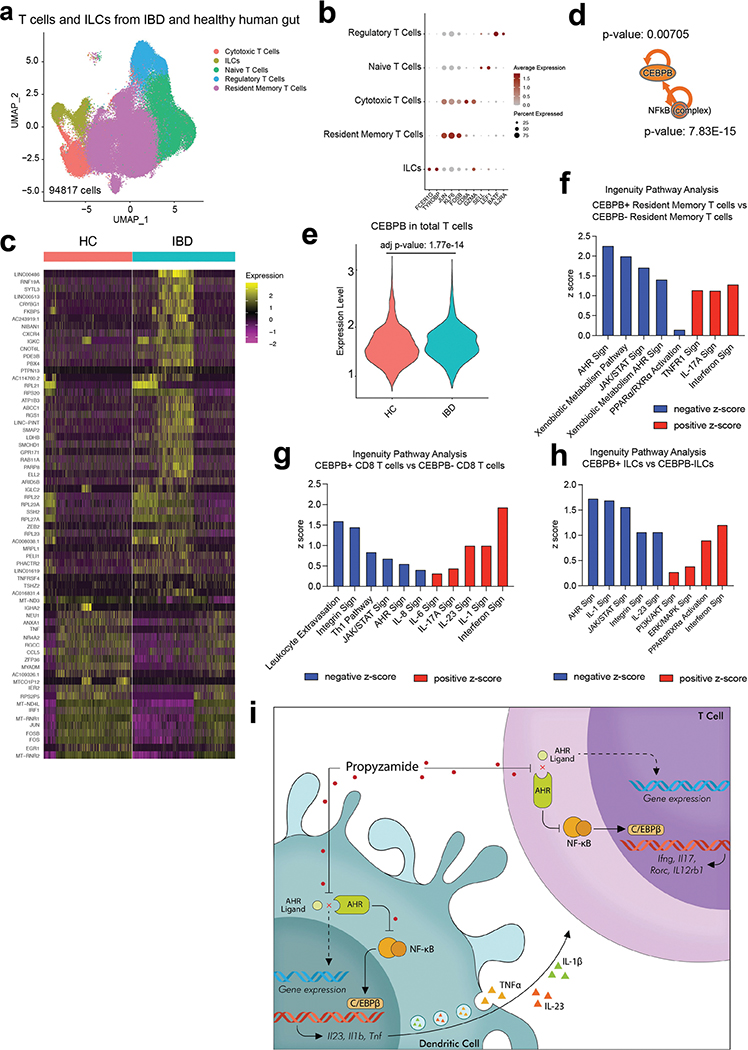
To evaluate the clinical relevance of these findings, we integrated three publicly available scRNA-seq databases26–28 corresponding to 58 patients with IBD and healthy control samples (Supplementary Table 9). DCs isolated from IBD lesions displayed an increased expression of pro-inflammatory genes, including those linked to NF-κB activation and CEBPB upregulation, particularly in the cDC1 subset (Extended Data Fig. 7e–k). Indeed, CEBPB+ DCs displayed increased expression of genes associated with NF-κB and other pro-inflammatory pathways, concomitant with decreased AHR signalling (Extended Data Fig. 7k). Together, these findings show that AHR–NF-κB–C/EBPβ signalling in DCs regulates TNBS-induced intestinal inflammation.
C/EBPβ signalling boosts colitogenic T cells
A network analysis of propyzamide-induced pathways in the EPA ToxCast database identified VCAM-1 as a potential target of propyzamide (Extended Data Fig. 8a). VCAM-1 mediates T cell adhesion to the vascular endothelium, and its expression in endothelial cells is induced in an NF-κB-dependent manner by pro-inflammatory cytokines such as TNF and IL-1β29. Indeed, propyzamide increased the transcriptional activity of the VCAM1 promoter in luciferase reporter assays, and upregulated Vcam1 expression in the colon of TNBS mice (Extended Data Fig. 8b,c). Propyzamide also increased T cell expression of the VCAM-1 ligand Itga4, but not Itga1, during TNBS-induced colitis (Extended Data Fig. 8d).
To investigate the role of VCAM-1 on the propyzamide-induced increase in T cell recruitment to the colon during TNBS-induced colitis, we used an anti-VCAM-1 blocking antibody (Extended Data Fig. 8e). VCAM-1 blockade abrogated the worsening of intestinal inflammation by propyzamide as determined by weight loss, colon shortening and histological analysis (Extended Data Fig. 8f–h). Indeed, VCAM-1 blockade also suppressed the recruitment of TH1 and TH17 cells to the colon (Extended Data Fig. 8i).
NF-κB activation boosts TH1 and TH17 cell differentiation30, but the role of C/EBPβ in this process is unclear. Propyzamide boosted TH1 and TH17 cell differentiation in vitro, as well as NF-κB activation (Extended Data Fig. 9a–f). Knockdown of Rela, encoding the p65 subunit of NF-κB, abrogated propyzamide-induced Cebpb upregulation in T cells, suggesting that NF-κB mediates the propyzamide-induced increase in Cebpb expression (Extended Data Fig. 9g, h). Indeed, after T cell treatment with propyzamide, C/EBPβ was recruited to the Ifng, Il12rb1 and Rorc promoters, and transactivated them in luciferase reporter assays (Fig. 4h,i). By contrast, knockdown of Cebpb decreased TH1/TH17 polarization and abrogated its boost by propyzamide in mouse (Fig. 4j and Extended Data Fig. 9i) and human T cells (Fig. 4k and Extended Data Fig. 9j), suggesting that C/EBPβ contributes to TH1 and TH17 cell differentiation.
To evaluate the role of C/EBPβ in the control of colitogenic T cells, we reconstituted RAG1-deficient mice with wild-type or C/EBPβ-deficient T cells, and induced colitis in the recipients with TNBS. C/EBPβ deficiency in T cells reduced weight loss, colon shortening and histopathology, as well as the number of colonic IFNγ+CD4+ and IL-17+CD4+ T cells (Fig. 4l–o), but did not affect ILCs or CD8+ T cells (Extended Data Fig. 9k).
Finally, the analysis of the scRNA-seq dataset of 58 patients with IBD and control individuals (Supplementary Table 9) detected NF-κB-driven C/EBPβ signalling concomitant with the upregulation in transcriptional modules associated to TH1 and TH17 cells in T cells from IBD lesions (Extended Data Fig. 10a–e). Interestingly, CEBPB+ resident memory T cells, cytotoxic T cells and ILCs in patients with IBD showed decreased AHR signalling (Extended Data Fig. 10f–h). Collectively, these findings show that NF-κB-driven C/EBPβ signalling promotes colitogenic T cell responses.
Discussion
Approximately 200 genetic loci have been associated with IBD1. However, genetic factors do not fully explain disease aetiology, and environmental exposures are thought to have a significant role in IBD onset and progression. Considering the multitude of exposures that affect humans in their lifetime, there is an unmet need for methods to evaluate the effects of the environment on IBD and other human disorders. Here we describe an integrated systems approach to identify environmental factors that promote intestinal inflammation and the mechanisms involved. The potential of zebrafish for microbiome research, and the reported effects of genomic31,32 and environmental33 factors on the microbiome, suggest that this approach may also be useful to study the effects of environmental factors on aspects of the microbiome relevant to IBD.
Our findings add to recent reports on the contribution of dietary emulsifiers15, oxazoles34 and other environmental factors to the pathogenesis of IBD and its increasing prevalence in industrialized nations. Propyzamide is broadly used for weed control during the production of vegetables, fruits and ornamental plants, and also in golf courses and sport fields. Although propyzamide is metabolized by plants, about 60% of the chemical remains unmetabolized 50 days after its application35. Indeed, propyzamide exposure levels have been estimated by the EPA to be 102 parts per billion (ppb) for surface water and 21 ppb for ground water for acute exposure, and 47 ppb for surface water and 18.6 ppb for ground water for chronic exposure36, similar to water surface concentrations of other environmental chemicals such as benzene, toluene diisocyanate and diethylhexyl phthalate, which have all been shown to have adverse health effects at exposure levels within this range37–42. Exposure levels to propyzamide would be expected to be much greater in communities in which higher levels of ground water contamination can occur from propyzamide use, as well as for people working in the agricultural sector who would be more directly exposed on a regular basis. Future studies should determine actual exposure levels in communities with potential high exposure to propyzamide and among agricultural workers, and determine whether other environmental, microbiome and genetic factors synergize with propyzamide during the pathogenesis of IBD.
AHR hyperactivation by dioxins has been linked to human pathology43, but these pathogenic effects involve slowly metabolized AHR agonists and not a rapidly metabolized molecule that interferes with AHR signalling like propyzamide. AHR participates in the control of intestinal inflammation and its regulation by the commensal flora32. Propyzamide boosts intestinal inflammation by interfering with the suppression by AHR of pro-inflammatory responses driven by NF-κB–C/EBPβ signalling. Thus, our findings suggest that propyzamide interferes with physiological AHR-dependent anti-inflammatory mechanisms that are activated by endogenous or microbial agonists. On the basis of the systemic distribution of propyzamide detected after oral administration, propyzamide may interfere with the physiological anti-inflammatory effects of endogenous or microbiome-produced AHR agonists in cells located in the intestine (such as intestinal epithelial cells, T cells and DCs), and also systemically. Notably, CARD9 polymorphisms linked to IBD promote dysbiosis, reducing the levels of microbiome-produced anti-inflammatory AHR agonists32. In this context, the recent description of AHR antagonists produced by the human microbiome44 suggests that the pro-inflammatory effects of propyzamide may be recapitulated, at least partially, by some microbial metabolites.
Propyzamide promotes microtubule disruption in plants21, an important point considering the reported link between microtubule stability and NF-κB activation22. However, propyzamide did not destabilize mammalian microtubules. Instead, our findings suggest that propyzamide releases NF-κB from its suppression by AHR. NF-κB has pivotal roles in inflammation, promoting TH1 and TH17 cell differentiation through its effects on antigen-presenting cells and T cells30,45–48. Indeed, we found that NF-κB activation in DCs and T cells promotes the expression of C/EBPβ, a transcription factor identified in genetic studies to contribute to IBD through unknown mechanisms49. Moreover, we established that NF-κB–C/EBPβ signalling promotes TH1 and TH17 cell differentiation by boosting the expression of T cell-polarizing cytokines in DCs, and also of cytokine receptors and other genes associated with T effector cells. Notably, these pro-inflammatory roles of NF-κB–C/EBPβ signalling were detected in samples from patients with IBD and TNBS-induced colitis in the absence of propyzamide administration, highlighting the power of integrated systems approaches to identify not only environmental exposures of interest, but also immunoregulatory pathways and mechanisms of disease pathogenesis. Moreover, NF-κB–C/EBPβ signalling may promote intestinal pathology through additional mechanisms, as suggested by reports linking C/EBPβ to the control of monocyte-induced fibrosis50, an important contributor to IBD pathology.
In summary, using an integrated systems approach in combination with machine learning, we identified an AHR–NF-κB–C/EBPβ pathway that regulates intestinal inflammation and is targeted by the common herbicide propyzamide (Extended Data Fig. 10i). Thus, the systematic investigation of environmental factors may define mechanisms of disease pathogenesis, guide epidemiological studies and identify candidate therapeutic targets in IBD and other diseases.
Methods
Animals and husbandry
AB wild-type, lck:gfp and rag1-mutant strains were obtained from the Zebrafish International Resource Center, bred and maintained at the aquatics facility at Brigham and Women’s Hospital. rag2-mutant strains were donated by L. Zon (Boston Children’s Hospital). The collection of embryos and maintenance of larvae were carried out as described previously5,8,51. Eight-week-old male C57BL/6J, B6(Cg)-Zbtb46tm1(HBEGF) Mnz/J, FVB.Cg-Tg(HIV-EGFP,luc)8Tsb/J, B6.D2N-Ahrd/J, B6.129S7-Rag1tm1Mom/J and B6.Cg-Tg(TcraTcrb)425Cbn/J mice were obtained from the Jackson Laboratory, and Cebpb+/+ and Cebpb−/− (Cebpbtm1Vpo/J) mice were obtained from A. Mildner. B6.129S4-Il17atm1.1Lky/J mice were a gift from T. Korn. Animals were kept in a pathogen-free facility at Hale Building for Transformative Medicine, Brigham and Women’s Hospital. Germ-free mice were bred in-house in tightly controlled and monitored isolators (Class Biologically Clean company) in an animal facility specifically dedicated to housing germ-free mice in the Massachusetts Host-Microbiome Center at the Brigham and Women’s Hospital. All of the experimental protocols were approved by the Institutional Animal Care and Use Committee of Brigham and Women’s Hospital.
Induction of intestinal inflammation in zebrafish
To induce intestinal inflammation, 7 d.p.f. zebrafish larvae were incubated in E3 medium52 containing 25 μg ml−1 TNBS (P2297, Sigma-Aldrich) for 24–72 h. For large-scale screenings, 7 d.p.f. zebrafish were placed in 48-well plates, containing a final volume of 600 μl, with 3 fish per well and at least 4 replicate wells per group. Candidate chemicals were diluted in DMSO to a stock concentration of 20 mM, and added to wells for a final concentration of 0.2, 1, 5 or 20 μM, as indicated in the figures. DMSO was used as a vehicle control in groups containing no candidate chemicals. Beginning at 7 d.p.f., zebrafish were fed every 24 h with 41.6 μg ml−1 Gemma Micro 75 food solution sterilized by autoclave (Ziegler). Note that, in each experiment, NOS inhibitor, (S-methyl-l-thiocitrulline acetate, Millipore-Sigma M5171) and FICZ (Tocris Bioscience, 5304) were used as positive controls to benchmark the results. Then, 24 or 72 h after induction, zebrafish were anaesthetized in E3 medium containing tricaine (MS-222, Sigma-Aldrich). Each zebrafish was transferred to a glass slide and manipulated using a pipette tip to lay on its side. Gut architecture was visualized and photographed using an inverted microscope (Zeiss) at ×10 magnification. The photos were blinded and scored for intestinal inflammation using the following 0–5 scoring system adapted from ref.8 as follows: 0, healthy narrow gut with good morphology and visible crypts and villi throughout; 1, slightly widened gut, good visible crypt morphology; 2, slightly widened gut, poor crypt morphology; 3, widened gut, little to no visible crypt morphology; 4, extremely widened gut, no visible crypts or villi; 5, death.
Chemicals
All chemicals were dissolved in DMSO at a stock concentration of 20 mM. Additional chemicals used in this study included S-methyl-l-thiocitrulline acetate (NOS inhibitor, Millipore-Sigma M5171), propyzamide (Millipore-Sigma, 45645); FICZ (Tocris Bioscience, 5304); lenaldekar53,54 (Millipore-Sigma, 5.31068). The NF-κB inhibitors used included Bay11–7082 (Millipore-Sigma, B5556) and Triptolide (Millipore-Sigma, T3652).
Machine learning-based chemical identification
The AC50 matrix of ToxCast data, which included information about 9,298 chemicals and 1,569 bioassays, was downloaded from the EPA website (https://www.epa.gov/chemical-research/exploring-toxcast-data-downloadable-data)55. An initial subset of 936 chemicals was analysed for their activity in 49 bioassays selected on the basis of their association with genes previously linked to IBD pathogenesis, including inflammatory cytokines such as TNF56, interferons57 and IL-1β58, and signalling molecules such as JAK/STAT59, PPAR60 and AHR61 (Supplementary Table 1). Of those 936 chemicals, 111 were selected for evaluation in the TNBS-induced model of intestinal inflammation in zebrafish on the basis of known human exposures and high chemical production volumes (Supplementary Table 3). On the basis of this in vivo screen of zebrafish, we defined 49 experimentally validated chemicals, which included 13 IBD-promoting chemicals, 4 IBD-ameliorating chemicals and 32 chemicals that showed no effect. Using ToxCast AC50 information on these 49 chemicals, we applied a machine-learning-based approach to identify additional chemicals predicted to worsen IBD. Specifically, we first filtered the remaining 9,249 chemicals and 1,569 bioassays in the dataset according to 3 criteria: (1) Chemicals that correlated (P < 0.05, Spearman correlation, false-discovery rate (FDR) < 0.2955) with the 49 experimentally validated chemicals were preselected for further analysis; this first step in the filtering process was designed to preselect chemicals that will best fit our model. (2) Bioassays with missing data for more than 75% of the experimentally validated chemicals were removed. (3) Chemicals/bioassays with more than 75% missing values in the ToxCast AC50 matrix were removed.
Using these criteria, the original AC50 matrix was reduced to a dataset of 297 chemicals and 233 bioassays, referred to as the testing set. Any missing values in the selected data were imputed using the k-nearest neighbour method (Euclidean distance, k = 5). Furthermore, to prevent unbalancing the groups, a singular value decomposition method was applied to identify 13 ‘no effect’ compounds that were most representative62, enabling us to define a training set composed of 13 IBD-promoting chemicals, 4 IBD-ameliorating chemicals and 13 chemicals that showed no effect. To select the bioassays most relevant for the three-class (IBD-promoting, IBD-ameliorating, IBD-no effect) classification, a Kruskal–Wallis test was performed to identify the bioassays that could group 13 IBD-promoting, 4 IBD-ameliorating and 13 IBD-no effect chemicals separately, leading to the identification of 16 significant bioassays (P < 0.05, FDR < 0.6610) (Supplementary Tables 4 and 5) referred to as the ‘IBD bioactivity features’. As this analysis step was used to identify potentially information-rich molecular features, unadjusted P values were ultimately used to avoid increasing type II errors63.
On the basis of the IBD bioactivity features of the 30 training set chemicals, a RF model was trained to identify candidate IBD-promoting chemicals in the test set64. To rank the predicted IBD-promoting chemicals, we generated a relevance score for each predicted chemical using a RWR algorithm65,66 based on the following formula:
where W is the row-normalized adjacency matrix (transition matrix) obtained by the concept of weighted gene co-expression analysis (WGCNA)67; P0 is the initial probability vector with m elements (the number of selected chemicals), in which the 13 validated IBD-promoting chemicals with value 1/13 and other chemicals with value 0; r is the restart probability, which was set to 0.3 in this study; Pt denotes the probability vector after t iterations, indicating the relevance score of the chemicals with respect to IBD-promoting. The iteration stops when ‖Pt+1 − Pt‖ < 10−6. On the basis of this analysis, chemicals with high relevance scores were considered candidate IBD-promoting chemicals and ranked (Supplementary Table 6). The top 20 predicted chemicals were validated TNBS-induced model of intestinal inflammation in zebrafish.
Network analysis
Using the ToxCast database of assays, we identified gene targets activated by propyzamide in biochemical and cell-based assays (https://comptox.epa.gov/dashboard/chemical/invitrodb/DTXSID2020420). We then used the web-based analysis tool NetworkAnalyst68 to visualize the network of genes controlled by propyzamide (Extended Data Fig. 8a) using the protein–protein interaction tool and IMEx Interactome.
Induction of colitis with TNBS in mice
Mice were shaved on the back right below the neck, painted with 1% TNBS mixed in acetone/olive oil solution for presensitization and colitis was induced with 2.5% TNBS in 50% ethanol by rectal injection 7 days after presensitization as described previously69.
Propyzamide administration
Vehicle (200 μl; corn oil) or 100 mg kg−1 propyzamide was intraperitoneally administered for 3 days until TNBS presensitization. Then, 1 day later, 200 μl of vehicle (corn oil) or propyzamide at 100 mg kg−1 was given by oral gavage for 6 consecutive days until TNBS rectal challenge. Mice were euthanized 3–5 days later for tissue collection.
VCAM1 blockade
Anti-mouse VCAM-1 antibody (100 μg; M/K-2.7, BioXcell) or isotype control antibody (rat IgG1, BioXcell) was administered intraperitoneally daily for 5 consecutive days, starting on the same day that mice received presensitization with TNBS.
Anti-CD3-antibody-induced enteritis
Vehicle (200 μl; corn oil) or 100 mg kg−1 propyzamide were administered by gavage for 7 days. Then, 200 μg of anti-CD3 monoclonal antibody (InVivoPlus; BP0001-1, Bio X Cell) were injected i.p. and mice were killed at the stated time points. To evaluate enteropooling, mice were fasted overnight but allowed to drink water ad libitum before anti-CD3 antibody injection, and euthanized 3 h after injection. The jejunum was then excised and carefully isolated after ligation at both ends, adherent mesentery was cut off, and jejunum segments were weighed, lengths were measured, and weight/length ratios were determined. To evaluate enteritis, the length of villi and the depth of crypts were measured, and the villus length/crypt depth ratio was calculated. The TUNEL assay was used to identify apoptotic cells in the jejunal tissue. In brief, jejunal samples that were previously fixed in Bouin’s solution and then embedded in paraffin were deparaffinized and rehydrated by subsequent incubations with xylene, 100% ethanol, 96% ethanol, 70% ethanol and PBS. TUNEL staining was performed according to the manufacturer’s instructions (11684795910, Sigma-Aldrich). Sections were analysed on the Leica DMi8 fluorescence microscope, and positive cells were quantified using ImageJ.
Histological analysis
Colon tissues were fixed in Bouin’s solution at 4 °C for 24 h and replace with 70% ethanol for long-term storage before histological process. Fixed samples were sent to Tufts or Harvard histology core facility for embedding, sectioning and H&E staining, and scored in a blinded manner as described previously9. Images were processed with ImageJ.
Isolation of colon-infiltrating cells
Mononuclear cells were isolated as described previously with minor modifications9. The colon was flushed, cut open longitudinally and incubated in 5 mM EDTA and 10% FBS-containing HBSS buffer to wash out intraepithelial mononuclear cells. The colon was then cut into small pieces, incubated in 5 ml digesting buffer contained 500 U collagenase from Clostridium histolyticum (C2139, Sigma-Aldrich), 2.5 μg DNase I (10104159001, Roche) and 10% FBS (Gibco) at 37 °C for 2 h. The digested colon was homogenized using a 70 μm strainer. CD45+ cells were isolated using a CD45 magnetic beads isolation kit (130-052-301, Miltenyi Biotec).
Flow cytometry
Cells were restimulated in DMEM containing 10% fetal bovine serum (Gibco), 500 ng ml−1 ionomycin (Sigma-Aldrich), 500 ng ml−1 PMA (Sigma-Aldrich) and BD GolgiStop (BD Bioscience), incubated at 37 °C and 5% CO2 for 4 h. Restimulated cells were then washed with PBS and stained for fluorescence-activated cell sorting (FACS) using the following antibodies: BUV661- or APC-labelled anti-CD45 (612975, BD Biosciences and 17-0451-83, eBioscience, 1:100), BV650- or BV421-labelled anti-CD3 (740530, BD Biosciences and 100227, BioLegend, 1:100), PE-Cy7-, BV605- or Alexa 700-labelled anti-CD4 (100422, 100547 and 100430, BioLegend, 1:100), BUV805-labelled anti-CD8a (564920, BD Biosciences), BV421-labelled anti-γδ (562892, BD Biosciences, 1:100), BV570-labelled anti-Ly6C (128030, BioLegend, 1:100), BV786-labelled anti-CD11b (101243, BioLegend, 1:100), FITC- or BV750-labelled anti-NK1.1 (108706, BioLegend and 746876, BD Biosciences, 1:100), BV711-labelled anti-CD127 (565490, BD Biosciences, 1:100). Cells were fixed using the FOXP3 staining buffer kit (Thermo Fisher Scientific), and intracellular staining was performed using PE-labelled anti-IL17a (12-7178-42, BioLegend, 1:100), APCCy7- or FITC-labelled anti-IFNγ (BD Biosciences, and 505806, BioLegend, 1:100), PECy7-labelled anti-hNGFR (345110, BioLegend, 1:100), APC-labelled anti-RORγt (17-6988-82, Invitrogen, 1:100), AF488-labelled anti-NF-κB p65 (4886S, Cell Signaling Technology, 1:50) antibodies. The samples were acquired using FACS DIVA in LSR-Fortessa or Symphony A5 (BD Bioscience) and analysed using FlowJo.
Pharmacokinetics
The plasma, faeces and urine pharmacokinetic parameters of propyzamide (100 mg kg−1) were determined in adult male C57BL/6 mice after a single oral administration. Propyzamide was solubilized in corn oil at 10 mg ml−1, and vortexed for 3 min before administration. The animals were subsequently observed for clinical signs, such as emesis, to ensure that the full oral dose was ingested. Mice were sampled at 0.5, 1, 2, 4, 8 and 24 h after dosing, semi-serial bleeding for plasma. Approximately 110 μl whole blood per time point was collected in K2 EDTA tube through the facial vein. Blood samples were put on ice and centrifuged for 5 min to obtain a plasma sample within 15 min. The mice were housed in metabolite cages. Faeces and urine samples were collected in fractions of 0–4 h, 4–8 h, 8–24 h after propyzamide administration. After collection, the weight of each faeces sample and the volume of each urine sample were recorded, and samples were conserved at −70 °C until bio-analysis.
qPCR
RNA was extracted using the RNAeasy kit (74106, Qiagen). For zebrafish experiments, two zebrafish from each well were combined and homogenized using a tissue homogenizer (THB115, Omni International) directly in RLT lysis buffer, then RNA was extracted using the RNAeasy kit (74106, Qiagen). cDNA was prepared using the High-Capacity cDNA Reverse Transcription Kit (43-688-13, Thermo Fisher Scientific). TaqMan probes and TaqMan Fast Universal PCR Master Mix (4444965, Thermo Fisher Scientific) were used. Target gene Ct values were normalized to eef1a1 or gapdh. For Fig. 1 and Extended Data Fig. 1, the ΔΔCt method of analysis was used, with values shown equal to where ΔΔCt = ΔCt (treated sample) − mean ΔCt of control samples. The following TaqMan probes were used in the study: zebrafish: eef1a1 (Dr03432748_m1) il1b (Dr03114368_m1), tnfa (Dr03126850_m1), ifng (Dr03081923_m1), il17a/f (Dr03096843_g1), nos2a (Dr03124734_m1); mouse: Actb (Mm02619580_g1), Tnf (Mm00443258_m1), Il10 (Mm00439614_m1), Il23 (Mm00518984_m1), Il1b (Mm00434228_m1), Ifng (Mm00801778_m1), Il17a (Mm00439618_m1), Csf2 (Mm01290062_m1), Tbx21 (Mm00450960_m1), Rorc (Mm01261019_g1), Vcam1 (Mm01320970_m1), Il12rb1 (Mm00434189_m1), Il6 (Mm00446190_m1), Tgfb1 (Mm01178820_m1), Cebpb (Mm00843434_s1), Rela (Mm00501346_m1), Cypa1a (Mm00487218_m1), Cyp1b1 (Mm00487229_m1) and Gapdh (Mm99999915_g1); human: CEBPB (Hs00270923_s1), GAPDH (Hs02786624_g1), IL23 (Hs00372324_m1), IL1B (Hs01555410_m1), TNF (Hs00174128_m1), CYP1A1 (Hs05011273_s1).
RNA-seq
Mice were euthanized 3 days after TNBS-colitis induction. Cells from the colon were isolated, lysed and RNA was isolated using the RNeasy Mini kit (74106, Qiagen) with on-column DNase I digestion (79254, Qiagen). RNA was suspended in 30 μl of nuclease-free water for sequencing using the 30 Digital Gene Expression70 by the Broad Technology Labs and the Broad Genomics Platform. Processed RNA-seq data were filtered, removing genes with low read counts. Read counts were normalized using TMM normalization and counts per million were calculated to create a matrix of normalized expression values.
RNA-seq data processing
The RNA-seq data were aligned to GRCm38 Mouse Genome using STAR (v.2.7.3a)71 and quantified using RSEM72 and Kallisto73. The results were then imported into R using tximport74. The differential expression analysis was performed using DESeq275 and the apeGLM algorithm76 was used to shrink the log2-transformed fold change for the purpose of accuracy. Heat maps were generated using the Gene-E program, and the z-scores were calculated for each gene-row using the mean expression of biological replicates. Data are row-centred, log2-transformed and saturated at levels −1.0 and +1.0 for visualization satisfying a FDR < 0.1.
Pathway and statistical analysis
Differential gene expression analysis results were used as input in IPA (https://digitalinsights.qiagen.com/IPA). Canonical and pathway analysis were performed to evaluate the activation/inhibition of pathways and upstream regulators using the log2-transformed fold changes of the corresponding targets. In summary, IPA has curated a database with the downstream targets of the upstream regulators, by matching the log2-transformed fold changes to the downstream regulators an activation score can be calculated, and the score is z-score-transformed by comparing it to all of the activation scores from other upstream regulators to determine activation and inhibition (https://digitalinsights.qiagen.com/IPA). Canonical pathways and upstream analysis metrics were considered to be significant at P < 0.05.
GSEA (Broad Institute, v.4.2.1) was used to generate enrichment plots for scRNA-seq data using Hallmark gene sets (h.all v.7.4) and Curated gene sets (c2.all v.7.4). In all cases, statistical analysis using GSEA or GSEAPreranked was determined using one-tailed t-tests in GSEA.
scRNA-seq of mouse colons
After cell isolation described above, the 3′ CellPlex Kit (10x Genomics, PN-1000261) was used for cell multiplexing according to the manufacturer’s protocol. After multiplexing, approximately 8,000–20,000 single cells (up to 42.4 μl) were loaded onto a single lane of Chromium Next GEM Chip G (10x Genomics, PN-2000177) and processed on the Chromium instrument (10x Genomics). Gene expression and multiplex libraries were prepared using the Single Cell 3′ Reagent Kits v.3.1 (10x Genomics) according to the manufacturer’s protocol. cDNA samples were amplified using the following PCR conditions: 98 °C (3 min); then 11–12 cycles of 98 °C (15 s), 63 °C (20 s), 72 °C (1 min); 72 °C (1 min); 4 °C hold. After PCR, cDNA samples were purified on the 10x Magnetic Separator (PN-230003) using the SPRIselect reagent (Beckman Coulter, B23317). cDNA was run on a Bioanalyzer High Sensitivity DNA chip (Agilent Technologies, 5067-4626) on the 2100 Bioanalyzer (Agilent). Then, 10 μl of cDNA was used for generating 3′ gene expression library and were indexed uniquely using the Dual Index Plate TT Set A (10x Genomics, PN-3000431). Samples underwent PCR using the following conditions: 98 °C (45 s); then 14 cycles of 98 °C (20 s), 54 °C (30 s), 72 °C (20 s); 72 °C (1 min); 4 °C hold. For generating cell multiplex library, 5 μl of cDNA were used and were indexed uniquely using the Dual Index Plate NN Set A (10x Genomics, PN-3000482). The samples underwent PCR using the following conditions: 98 °C (45 s); then 6 cycles of 98 °C (20 s), 54 °C (30 s), 72 °C (20 s); 72 °C (1 m); 4 °C hold. Libraries were purified using SPRIselect reagent. The samples were then run on a 2100 Bioanalyzer to assess library size and were quantified by qPCR using the Library Quantification Kit (Kapa Biosystems, KK4824). Libraries were pooled and sequenced on the NovaSeq 6000 system (100 cycles) at an average depth of 20,000 reads per cell.
scRNA-seq analysis
The raw scRNA-seq data were downloaded from three published data repositories: Gene Expression Omnibus (GEO) GSE134809 and GSE134809 and the Gut atlas (https://gutcellatlas.org). The fastq files were processed using CellRanger Count using the default settings. For newly generated scRNA-seq data, the multiplexed single-cell dataset was processed using the CellRanger Pipeline ‘multi’ function to demultiplex and quantify samples77. The filtered count matrix generated by CellRanger was used in the downstream analysis. Scrublet78 was used to detect doublets in each sample. After detecting and removing doublets, cells in which we detected <500 or >2,500 genes, >10,000 transcripts, or >25% mitochondrial content were filtered out before the analysis.
Data were normalized using the negative binomial regression model with the top 4,000 variable genes, and the percentage of mitochondrial contents was regressed out in the normalization and scaling process79. Principal component analysis was performed, and the combination of canonical correlation analysis and reciprocal principal analysis was used to remove the batch effect80. The top 50 principal components were used to generate the UMAP and the clustering results using Seurat80.
The following canonical cell markers were used to call the cell type: CLEC10A, CD1C, FCER1A and FCER1G for DCs; CD3D, CD3E and CD3G for T cells. To cluster subsets, each population was renormalized using the log-normalization method80 and batch effects were removed using Harmony81. The first 50 principal components were used to conduct dimension reduction and clustering. For the T cell population, canonical markers such as JUN, KLF6, FOSB, CD8A, GZMA, SELL, LEF1, BATF and IL2RA were used to find subsets; for the DC population, BATF3, CLEC9A, CADM1, CLEC10A and CD1C were used to define subsets. To study the activation of AHR signalling under different conditions, the AHR-signalling pathway gene set was downloaded from the Gene Ontology Database82, and the module signature score of the AHR-signalling pathway was calculated using AddModuleScore in Seurat80. The signature score analysis was then performed using the Wilcoxon ranked-sum test. All of the downstream differential expression analyses were conducted using MAST83.
ChIP–seq data analysis
ChiP–seq peaks files were downloaded from the GEO repository (GSE36099) and the Integrated Genome Viewer (Broad Institute, v.2.11.3) was used to visualize peaks at selected loci.
Gut microbiome 16S sequencing
Faecal samples were collected from the ileum, jejunum, caecum and colon at the end of the study. DNA was extracted using the DNeasy PowerLyzer PowerSoil kit (12855, Qiagen) according to the manufacturer’s instructions. 16S rRNA gene V45 region was amplified and by PCR using HotMaster Taq DNA Polymerase and Hotmastermix (10847-708, VWR) and barcoded 515F fusion primers and reverse 926R fusion primers that contain adaptors for MiSeq sequencing and single index barcodes so that the PCR products can be pooled. These primers were designed by the Earth Microbiome Project to amplify most bacteria84,85. DNA was then quantified using the Quant-iT PicoGreen dsDNA Assay Kit (P11496, Thermo Fisher Scientific) and 100 ng of each sample were pooled and cleaned-up using the QIAquick PCR Purification Kit (28104, Qiagen). DNA was requantified after clean-up using the Qubit Fluorometric Quantification kit (Thermo Fisher Scientific) and submitted for paired-end 300 bp read sequencing on the Illumina MiSeq instrument at the Harvard Medical School Biopolymers Facility as described previously86. Quantitative insights for microbial ecology software 2 (QIIME2) was used for quality filtering and downstream analysis for alpha and beta diversity, and compositional analysis according to standardized protocols87. Dada2 was used to trim the forward read to 220 bp and the reverse reads to 190 bp, and to denoise, quality filter sequences, join reads and dereplicate the reads into amplicon sequence variants. Taxonomic assignment was performed using the RDP classifier built into QIIME2, which was trained against the 16S rRNA V45 region database from EZBioCloud database88. Distances between samples (β-diversity), were calculated using the phylogenetic-based distance UniFrac89. Statistical testing for differential clustering of samples on the PCoA plots was performed using the PERMANOVA test using 999 permutations.
Faecal microbiota transplant
Faecal microbiota was isolated from propyzamide- or vehicle-treated mice. To eliminate the potential contamination with propyzamide, propyzamide treatment was stopped 3 days after TNBS induction and fresh faeces was collected from day 6–11 after TNBS colitis induction. Germ-free mice were orally gavaged for 5 consecutive days with 200 μl of a freshly prepared faecal slurry containing 8 × 108 CFU per ml of bacteria as we described previously17. Reconstitution efficiency was assessed 2 days after the last transplant by qPCR analysis of bacterial 16S rRNA as described previously90. In brief, DNA was isolated from faeces through purification using a DNeasy PowerSoil Kit (Qiagen, 12888-100) and DNA was quantified using the Quant-iT dsDNA Assay Kit, high sensitivity (Thermo Fisher Scientific, Q33120). Normalized amounts of DNA were used as input for qPCR of 16S rRNA levels using the Fast SYBR Green Master Mix (Thermo Fisher Scientific, 4385612). The following 16S rRNA primers were used UniF340, 5′-ACTCCTACGGGAGGCAGCAGT-3′; and UniR514, 5′-ATTACCGCGGCTGCTGGC-3′. Downstream analyses were performed 10 days after reconstitution.
Study participants and blood collection
Healthy peripheral blood mononuclear cell (PBMC) donors were recruited at Brigham and Women’s Hospital. All of the procedures were approved by the Institutional Review Board of Brigham and Women’s Hospital and informed consent was obtained from each participant. Exclusion criteria included pregnancy, history of gastrointestinal surgery, intestinal bowel disease, and antibiotics or vaccines within the past 3 months. Blood PBMCs were isolated by Ficoll-Paque PLUS density gradient (Sigma Aldrich, GE17-1440-03).
Human DC and T cell cultures
DCs were enriched from PBMCs using the Pan-DC enrichment kit (Miltenyi, 130-100-777), and enriched DCs were stained with BV421 anti-human CD11c (BioLegend, 301628, 5 μl per sample), APC Cy7 anti-human HLA-DR (BioLegend, 307618, 5 μl per sample), FITC anti-human CD3 (BD Biosciences, 555232, 10 μl per sample) and PE anti-human CD123 (BD Biosciences, 340545, 5 μl per sample) antibodies. T cells were sorted from the non-DC fraction as CD3+ cells. DCs sorted as CD3−CD11c+HLA-DR+CD123− were silenced and 48 h cells were stimulated with LPS (100 ng ml−1) and 10 μg ml−1 of OVA 323-339 (SP-51023-1, Genemed) in the presence of propyzamide (10 μM) or vehicle. After 6 h, cells were washed and co-cultured with allogenic T cells at a 1:10 ratio for 48 h. Cytokines were quantified in the supernatants using the LEGENDplex Th cytokine panel (BioLegend, 741027). CEBPB was silenced on T cells using the Human T cell nucleofection kit (Lonza, VPA-1002) and the Accell Human CEBPB-siRNA smart pool (Dharmacon, E-006423-00-0005). T cells were then activated by plate-bound anti-human CD3 (Thermo Fisher Scientific, 16-0037-85) and soluble anti-human CD28 (Thermo Fisher Scientific, 16-0289-85) in presence of propyzamide (10 μM) or vehicle.
Luciferase assay
HEK293FT or DC2.4 (SCC142, Millipore) cells were cultured in DMEM supplied with 10% FBS (GIBCO) and penicillin–streptomycin (Thermo Fisher Scientific). VCAM1, Il1b, Tnf, Il23, Ppara, Rara, IFNG, RORC and IL12RB promoter reporter plasmids expressing gaussia luciferase were acquired from GeneCopoeia; pGud-Luc has been described previously91,92. Reporters were transfected with Lipofectamine 2000 (11668019, Thermo Fisher Scientific). To evaluate AHR promoter activity, cells were stimulated with vehicle, propyzamide (10 μM), FICZ (0.5 μM) or a combination of propyzamide and FICZ. To evaluate VCAM1 promoter activity, cells were stimulated with vehicle or propyzamide for 2 more days before medium collection. CEBPB plasmid (15738, Addgene) or pGL4.54[luc2/TK] vector (E506A, Promega) were co-transfected to evaluate C/EBPβ binding to the Il1b, Tnf, Il23, RORC, IL12RB1 or IFNG promoters. Luciferase activity was evaluated using the Gaussia Luciferase Flash Assay Kit (16159, Thermo Fisher Scientific).
AHR competitive binding assay
Human and mouse AHR proteins were synthesized using AHR expression constructs (pSporthAHR2 and pSportmAHR, respectively; from C. A. Bradfield)93,94 using the TnT-Quick Coupled Reticulocyte Lysate System (SP6; Promega). Competition with 2,3,7,8-tetrachloro[1,6–3H] dibenzo-p-dioxin ([3H]TCDD; 27.5 Ci mmol−1; Chemsyn Science Laboratories) for binding to AHR proteins was measured by velocity sedimentation on sucrose gradients in a vertical tube rotor, as described previously95,96. Single TnT reactions were diluted 1:3 (human AHR) or 1:7 (mouse AHR) with MEEDMG buffer (25 mM MOPS, 1 mM EDTA, 5 mM EGTA, 0.02% NaN3, 20 mM Na2MoO4, 10% (v/v) glycerol, 1 mM DTT, pH 7.5) with protease inhibitors (Sigma-Aldrich, P8340 cocktail; 104 mM AEBSF [4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride], 80 μM aprotinin, 1 mM bestatin, 1.4 mM E-64 [N-(trans-epoxysuccinyl)-l-leucine 4-guanidinobutylamide], 2 mM leupeptin, 1.5 mM pepstatin, Sigma-Aldrich) and incubated overnight at 4 °C with [3H]TCDD (1.5–2.0 nM) ± propyzamide (Sigma-Aldrich, 45645; 300 or 1,000 μM final concentration, dissolved in DMSO) or CH223191 (100 μM final concentration; synthesized as described previously97). A 5 μl aliquot of each incubation was taken to determine the total concentration of [3H]TCDD and the remaining incubation was applied to 10–30% sucrose gradients. [14C]Catalase was included in each tube as a sedimentation marker. The samples were centrifuged for 140 min at 60,000 rpm at 4 °C in the VTi65.2 rotor. The fractions were collected and radioactivity in each fraction was determined by liquid scintillation counting. Non-specific binding was determined using TnT reactions without the AHR expression vector. Specific binding was measured as the sum of radioactivity in the AHR peak, after subtracting non-specific binding. In each experiment, results were normalized to the amount of specific binding in the absence of competitor.
Isolation of mouse splenocytes
Spleens were isolated and mechanically dissociated. Red blood cells were lysed with ACK lysing buffer (A10492-01, Life Technology,) for 5 min and washed with 0.5% BSA, 2 mM EDTA pH 8.0 in 1× PBS and prepared for downstream applications.
Sorting of mouse DCs and T cells
DCs were sorted after isolation from spleen or colon from propyzamide- or vehicle-treated mice as described above. Cell suspensions were stained with FITC anti-mouse-NK1.1 (108706, BioLegend), CD64 (139316, BioLegend), Ly6G (127606, BioLegend), Ly6C (128006, BioLegend), B220 (553087, BD Bioscience) F/480 (11-4801-85, Thermo Fisher Scientific), PECy7 or PE-Dazzle594 anti-mouse-CD11c (N418, BioLegend) or PECy7-anti mouse CD3 (100220 and 117348, BioLegend) antibodies. After 30 min on ice, cells were washed and sorted on the FACS Aria Ilu (BD Biosciences) system. T cells were used to reconstituted Rag1-deficient mice as described below. DCs were pulsed with 10 μg ml−1 of OVA 323-339 (SP-51023-1, Genemed) for 6 h, washed and co-cultured with naive CD4+ T cells isolated from spleens from B6.Cg-Tg(TcraTcrb)425Cbn/J OT-II mice using magnetic beads (130-104-453, Miltenyi). After 48 h, cells were restimulated and stained as described below.
NF-κB activation assay
Splenic DCs isolated from FVB.Cg-Tg(HIV-EGFP,luc)8Tsb/J reporter mice using magnetic beads (130-125-835, Miltenyi) were stimulated for 1 h with test chemicals at a final concentration of 10 μM. Fluorescence was determined by flow cytometry.
BMDCs
To obtain bone marrow progenitors, the femur and tibiae from C57BL/6J wild-type mice were removed and cells were flushed out with 1 ml of 0.5% BSA, 2 mM EDTA pH 8.0 in 1× PBS. Red blood cells were lysed with ACK lysing buffer (Life Technology, A10492-01) for 5 min and washed with 0.5% BSA, 2 mM EDTA pH 8.0 in 1× PBS. For DC differentiation, cells were cultured in Petri dishes (100 mm) at a density of 1 × 106 cells per ml in DMEM/F12 + GlutaMAX supplemented with 10% FBS (10438026, Life Technologies,) and 1% penicillin–streptomycin (15140122, Life Technologies) containing 20 ng ml−1 of GM-CSF (315-03, Preprotech). The medium was replaced at days 3 and 5. Cells were collected for downstream applications at day 8. BMDCs were activated with 100 ng ml−1 Ultrapure LPS, E. coli 0111:B4 (InvivoGen, tlrl-3pelps) in the presence of DMSO (vehicle) or propyzamide (2 μM for 6 h.
T cell activation in vitro
For in vitro experiments, splenic naive CD4+ T cells were isolated from C57BL/6J wild-type mice using magnetic beads (130-104-453, Miltenyi) and activated with anti-CD3 (10 μg ml−1, BE0001-1-A005mg, BioXcell) and anti-CD28 (0.25 μg ml−1, BE0015-5-A005mg, BioXcell) antibodies in the presence of DMSO (vehicle) or propyzamide (2 μM). Cells were polarized as we previously described98. TH1 cells were generated with IL-12 (30 ng ml−1, 419-ML-010/CF, R&D Systems) and TH17 were induced by IL-6 (30 ng ml−1, 406-ML-005, R&D Systems) and TGFβ1 (3 ng ml−1, 130-095-067, Miltenyi) stimulation for 48–72 h.
RNA silencing of DCs
A siRNA pool (1.5 μl of 20 μM) was mixed with 2 μl interferin (409-10, Polyplus-transfection) in 100 μl Opti-MEM (31985062, Life Technologies). The mix was incubated for 10 min at room temperature and added to 500 μl of complete DMEM on a 24-well plate. Then, 48 h later, cells were used for downstream assays. The siRNA pools used were siRelA (L-040776-00-0005, Dharmacon), siCebpb (L-043110-00-0005, Dharmacon), siCEBPB (E-006423-00-0005, Dharmacon) and siNon-targeting (NT) (D-001810-10-20, Dharmacon).
RNA silencing of T cells
Cebpb or Rela expression was silenced on naive CD4+ T cells using the Mouse T Cell Nucleofector Kit (VPA-1006, Lonza). In brief, 1 × 106 naive CD4+ T cells were resuspended in 100 μl of room temperature nucleofector solution containing 30 pmol of Rela siRNA (E-040776-00-0005, Dharmacon), Cebpb siRNA (E-043110-00-0005, Dharmacon) or non-targeting siRNA (D-001910-10-05, Dharmacon) per sample. Cell suspensions were transferred to a cuvette, placed onto cuvette holder and nucleofected using the X-001 program of the Nucleofector II device (AAB-1001, Lonza). Next, 500 μl of pre-equilibrated complete medium was added to cell suspensions and then transferred to 1.5 ml pre-equilibrated medium on a 12-well plate. After 3 h, cells were plated on plates that were precoated anti-CD3 antibodies with soluble anti-CD28 antibodies and propyzamide (2 μM) or vehicle. After 48 h, cells were frozen in RLT buffer for RNA extraction.
DC and T cell co-culture
BMDCs previously silenced as described above or primary DCs from Cebpb-deficient mice were stimulated with LPS (100 ng ml−1) plus OVA peptide (ISQAVHAAHAEINEAGR) (10 μg ml−1) and vehicle or propyzamide (2 μM) overnight. Naive CD4+ T cells were isolated from spleens from B6.Cg-Tg(TcraTcrb)425Cbn/J OT-II mice using magnetic beads and co-culture at 1:10 ratio (BMDCs:T cells). After 48 h, the supernatants were collected for ELISA and RNA was isolated from the cells.
In silico promoter analysis
The Mus musculus Il23, Rorc, Il12rb1 and Ifng genomic sequences were obtained using Ensembl. The DNA sequences ~1,000 bp upstream and ~1,000 bp downstream of the protein-coding transcripts for Il1b, Il23, Tnf, Rorc, Il12rb1 and Ifng were analysed. CEBPB DNA-binding sites were defined using Mulan.
ChIP
BMDCs or T cells activated as described above were cultured with propyzamide or vehicle. After 48 h, cells were prepared according to the ChIP-IT Express Enzymatic Shearing and ChIP protocol (53009, Active Motif). In brief, cells were fixed in 1% formaldehyde, washed in PBS and glycine Stop-Fix solution. Cells were pelleted, and chromatin was sheared using the Enzymatic Shearing Cocktail (Active Motif) for 10 min at 37 °C. Sheared chromatin was immunoprecipitated with 5 μg of anti-C/EBPβ antibody (ab15050, Abcam) or mouse IgG control (ab37355, Abcam) overnight at 4 °C with rotation. The next day, magnetic beads were washed, and cross-links were reversed in 0.1% SDS and 300 mM NaCl TE buffer at 63 °C for 4 h. DNA fragments were purified using the QIAquick PCR Purification Kit (28104, Qiagen). qPCR was performed using the Fast SYBR Green Master Mix (4385612, Thermo Fisher Scientific). The following primer pairs were used: CEBP Il23 site 1 F, CATGACACGGGAACCAGACT; R, AGGGGCAGGGAAGTAATGGA; CEBP Il23 site 2 F, AACTTTTGAGAGCCTGCCGT; R, GTACAGCGATGATGACCCGT; CEBPB Rorc F, CGAAGCTCCCCAGCTAGAAC; R, GGGGTTTAAGCTCTGCTCCA; CEBPB Il12rb1 F, CCTTCAGCCCTGCAGAAGTT; R. GGCCACAAGGACAAAGAGGA; CEBPB Ifng F, GAGAGCCCAAGGAGTCGAA; R, TACCTGATCGAAGGCTCCTC; CEBPB Tnf F, GTGGAGAAAGACGGGGATG; R, ATCTGCTTGTTCATTCATTCATTC; CEBPB Il1b F, TCTCTTTATCTGGGGTGTGAGTT; R, AGCCCTCAGGTAGAGGAACC.
C/EBPβ DNA-binding ELISA
For C/EBPβ quantification, the Mouse/Human C/EBPβ DNA Binding Elisa Kit (LS-F816, LS Bio) was used. T cells stimulated under TH17-polarizing conditions and BMDCs were transfected with siRNA as described above in the presence of vehicle (DMSO) or propyzamide (2 μM) for 48 h, and nuclear extraction was performed according to the manufacturer’s instructions. Nuclear lysates (5 μg) and nuclear lysate positive controls were plated in duplicate in a 96-well microplate coated with streptavidin bound to biotinylated oligonucleotides and incubated on orbital shaker at room temperature for 2 h. The plate was washed three times with 1× wash buffer with gentle shaking in-between. Next, samples and nuclear lysate positive controls were incubated or not with primary antibody for antibody negative control and left on orbital shaker at room temperature for 2 h. The plate was washed three times with 1× wash buffer with gentle shaking in-between and incubated with HRP-conjugated anti-rabbit IgG antibody on an orbital shaker at room temperature for 1 h. The plate was washed three times with 1× wash buffer with gentle shaking in-between and revealed using 1× TMB substrate solution for 10 to 30 min. The reaction was stopped by stop solution and plate was read at 450 nm and 540 nm on the GloMax Explorer Multimode Microplate Reader (Promega). Correction was performed by subtracting reading at 540 nm from reading at 450 nm. Next, the reading of the primary antibody negative control was subtracted from the reading with the primary antibody, to correct for background noise. For the qualitative analysis, the relative sample concentration was determined by normalizing to siNT cells treated with vehicle.
Subcellular fractionation and immunoblot analysis
Nuclear protein was extracted using the Cell Fractionation kit (9038S, Cell Signaling) and 10 μg of nuclear fractions were separated by 4–12% Bis-Tris Nupage gels (Invitrogen) and transferred onto 0.45 μm PVDF membranes (Millipore). As primary antibodies, rabbit anti-lamin B monoclonal antibodies conjugated to HRP (D9V6H, Cell Signaling) and anti-NF-κB p65 rabbit monoclonal antibodies (D14E12, Cell Signaling) were used, followed by goat anti-rabbit IgG HRP-linked antibodies (7074S, Cell Signaling). Blots were developed using the Ultra Digital-ECL Substrate Solution (Kindle Bioscience), the signal was obtained using KwikQuant Imager (Kindle Bioscience) and the image was analysed using ImageJ.
Transduction of cumate-inducible C/EBPβ-containing lentivirus
A custom SparQ cumate-inducible lentiviral construct containing the mouse Cebpb gene (pCDH-CuO-MCS-IRES-GFP-EF1α-CymR-T2A-Puro, System Biosciences, QM812B-1) was transfected into HEK293FT cells according to the ViraPower Lentiviral Packaging Mix protocol (Thermo Fisher Scientific, K497500) and pseudotyped with pLP/VSVG. The medium was changed the next day, and the virus-containing medium was collected 48 h later, centrifuged and the supernatant was filtered through a 0.45 μm PVDF filter and frozen at −80 °C until use. Lentiviral transduction was performed using a modified Spinfection protocol99. A total of 250,000 freshly magnetically isolated (MACS Miltenyi, 130-125-835) primary splenic DCs were added to 0 or 125 μl lentiviral supernatant, with 8 μg ml−1 polybrene (EMD Millipore, TR-1003-G) in a 48-well plate and centrifuged at 2,000 rpm at 37 °C for 2 h. The medium was exchanged and the cells were left to rest overnight. The next day, 0, 10 or 50 μg ml−1 cumate solution (System Bioscience) was added to wells. Vehicle-only cells were pulsed with the highest-used volume of ethanol (cumate solution vehicle). Fluorescence microscopy was used to confirm expression. Cells were subsequently treated with 100 ng ml−1 lipopolysaccharide (Invivogen, tlrl-3pelps) for 6 h.
Bone marrow chimeras for the analysis of cDCs
Recipient mice were lethally irradiated with a dose of 9.5 Gy and 1 day later were given an intravenous injection of 5 × 106 bone marrow cells isolated from the femora and tibiae of B6(Cg)-Zbtb46tm1(HBEGF)Mnz/J mice. Recipients of bone marrow were then allowed to rest for 8 weeks before use. Bone marrow chimeras were inoculated intraperitoneally every other day for 2 weeks with 20 ng DTx per g body weight. Four to five randomly assigned mice were used per experimental group per experiment. Pre-DCs from wild-type or Cebpb-deficient mice were sorted using Alexa 488 anti-mouse CX3CR1 (149022, BioLegend), PE-Dazzle594 anti-mouse CD11c, PE anti-mouse CD135 (135306, BioLegend), FITC anti-mouse B220, APC-Cy7 anti-mouse MHC-II (307618, BioLegend) antibodies.
T cell transfer
Rag1-deficient mice were reconstituted with 500,000 splenic T cells from wild-type or Cebpb-deficient mice intraperitoneally. Four weeks after, T cell reconstitution was confirmed in peripheral blood and TNBS colitis was induced as described below.
Statistical analysis
Statistical analyses were performed using Prism software (GraphPad) using unpaired t-tests. P < 0.05 was considered to be significant and is indicated in the figures. All error bars represent the s.e.m.
Extended Data
Extended Data Fig. 1 |. TNBS-induced intestinal inflammation in Zebrafish.
(a) Zebrafish survival when treated with 25, 50, or 75 μg ml−1 TNBS for 24, 48, 72, or 96 h. Data shown as mean percent survival ±SEM of 3 independent experiments with n = 20 fish per group per experiment. (b) Intestinal scores of naive 10 d.p.f. zebrafish, or zebrafish exposed for 24 or 72 h starting at 7 d.p.f. to TNBS (25 μg ml−1). (n = 24 per group). (c) Intestinal scores of zebrafish exposed for 72 h starting at 7 d.p.f. to TNBS plus vehicle, NOS inhibitor (NOSi, 10 μM), or FICZ (10 μM). (n = 24 per group). (d–f) Intestinal scores (d) and lck (e), il17a/f, tnfa, ifng, il1b and nos2a (f) expression in 10 d.p.f. naive, or TNBS-exposed (25 μg ml−1, 72 h) vehicle- or lenaldekar-treated (LDK, 5 μM) zebrafish (n = 24 per group for intestinal scores, n = 6 for lck naive vehicle, n = 11 otherwise). (g) Intestinal scores in 10 d.p.f. naive vehicle or TNBS-exposed (25 μg ml−1, 72 h) WT, rag1- and rag2-deficient zebrafish. (n = 24 for WT naive and TNBS, n = 44 for rag1 and rag2 KO groups). (h–i) lck (h), il17a/f, tnfa, ifng, il1b and nos2a (i) expression in 10 d.p.f. TNBS-exposed (25 μg ml−1, 72 h) WT or rag2-deficient zebrafish. (n = 18 per group). (j) T cells expressing GFP under the control of the lck promoter in the intestine of naive 10 d.p.f. lck:gfp zebrafish, or lck:gfp zebrafish exposed for 72 h starting at 7 d.p.f. to TNBS (25 μg ml−1) alone or in combination with lenaldekar (LDK, 5 μM) or propyzamide (20 μM). Top panels are brightfield, middle panels are lck:gfp expression, bottom panels are a composite of both. (k) Quantification of intestinal GFP-positive T cells in lck:gfp zebrafish shown in (j). (n = 12 per group). (l) Activity of candidate chemicals in 49 ToxCast bioassays targeting genes linked to IBD; shown as active (red), inactive (blue), or no data (grey). (m) Principal component analysis based on ToxCast bioassays in chemicals found to ameliorate, promote, or have no effect on intestinal inflammation in the TNBS-induced zebrafish model. (n) Intestinal scores in zebrafish exposed for 72 h to TNBS and chemicals randomly selected from the ToxCast database at 0.2, 1, 5, or 20 μM. Each panel represents the average of n = 12 zebrafish per group. Grey panels indicate concentrations lethal to zebrafish larvae. (o,p) Intestinal scores (o) and il17a/f, tnfa, ifng, il1b and nos2a expression (p) in naive zebrafish treated with vehicle or propyzamide (0.2, 1, 5, or 20 μM) (n = 24 per group for intestinal scores, n = 9 per group for gene expression). (q) Intestinal scores of TNBS-exposed (25 μg ml−1, 72 h) zebrafish treated with vehicle, NOS inhibitor (10 μM), FICZ (10 μM), or propyzamide (20 μM), or a combination as indicated. (n = 36 for TNBS+vehicle and TNBS+propyzamide groups, n = 24 otherwise). Two-way ANOVA followed by Šídák’s multiple comparisons test for b. One-way ANOVA followed by Šídák’s or Dunnett’s multiple comparisons test for c–g,k,n,o,q. Unpaired student’s T test for h,i. Data shown as mean±SEM.
Extended Data Fig. 2 |. Propyzamide boosts TNBS-induced colitis in mice.
(a) Gating strategy used to analyse CD4+ T cells. (b) CD3+ lymphocytes in colon normalized by tissue length from vehicle- or propyzamide-treated (100 mg kg−1) mice during TNBS-induced colitis (n = 5 mice per group). (c) Representative dot plots of IFNγ and IL17 expression in CD4+ T cells. (d) Representative dot plots of IL-17 and RORγt expression in CD4 T cells and number of IL17+RORγt+ CD4 T cells in vehicle- or propyzamide-treated TNBS mice (n = 4 for vehicle, n = 3 for propyzamide). Unpaired student’s T test. (e) Ifng and Il17 expression determined by qPCR in lamina propria mononuclear cells (LPMC) from naive mice (n = 5) and vehicle- or propyzamide-treated TNBS mice (n = 4 mice per group). (f) IL17+γδ+ or CD8+ T cells isolated from colons of vehicle- (n = 7) or propyzamide-treated (n = 9) mice during TNBS-induced colitis. (g,h) Weight change (g) and colon length (h) of mice treated with vehicle or propyzamide (100 mg kg−1) for 10 days (n = 20 mice per group). (i) Total CD4+ (n = 10 propyzamide, n = 9 vehicle), IFNγ+ CD4+ (n = 17 per group) and IL-17+ CD4+ (n = 9 per group) T cells in colons of vehicle- or propyzamide-treated mice. (j) CD8+and IFNγ+ CD8+ T cells in colons of vehicle- (n = 9 for CD8+, n = 10 for IFNγ+ CD8+) or propyzamide-treated (n = 8 per group) mice. (k) γδ+T and IL-17+ γδ+T cells in colons of vehicle- or propyzamide-treated mice (n = 10 per group). (l) CD127+ ILCs (n = 10 vehicle, n = 7 propyzamide) and IL-17+ILC3s (n = 6 per group) in colons of vehicle- or propyzamide-treated mice. (m) Rela and Cebpb expression determined by qPCR from colonic CD45+ cells isolated from vehicle- or propyzamide-treated mice (n = 4 for propyzamide Rela, n = 5 otherwise). (n) Propyzamide concentrations in plasma (n = 6 per timepoint), faeces (n = 1) and urine (n = 1) after propyzamide administration (100 mg kg−1). (o) Propyzamide levels in plasma, faeces and urine collected from naive or TNBS-induced colitis mice (n = 3 per group). One-way ANOVA followed by Holm-Šídák’s or Tukey’s of multiple comparisons test for b and e. Data shown as mean±SEM.
Extended Data Fig. 3 |. Effects of propyzamide on the gut microbiome.
(a) α-diversity of the faecal microbiome (n = 6 for Jejunum vehicle, n = 7 for Ileum, Caecum and Colon vehicle, n = 8 for Ileum Propyzamide, n = 9 for Jejunum, Caecum and Colon propyzamide). Kruskal–Wallis nonparametric ANOVA test. (b) β-diversity shown as Principal-coordinate analysis (PCoA) based on unweighted UniFrac metrics. (c) Relative abundance of bacteria classified at a family-level taxonomy. (d) Relative abundance of the Suterellaceae family (n = 7 for TNBS vehicle, n = 9 for TNBS propyzamide, n = 10 for naive vehicle and propyzamide) Kruskal–Wallis nonparametric ANOVA test. (e) Schematic of faecal microbiota transplant (FMT) to germ free mice. This schematic was created using BioRender. (f) 16S quantification by qPCR after FMT (n = 4 control, n = 20 before reconstitution, n = 8 for FMT from vehicle- and propyzamide-treated mice). (g,h,i) Weight loss (g) (n = 10 vehicle, n = 6 propyzamide), colon length (h) (n = 10 vehicle, n = 7 propyzamide)and representative hematoxylin & eosin staining in colons (i) from germ free mice after FMT from propyzamide- or vehicle-treated mice (n = 8 vehicle, n = 5 propyzamide for quantification). (j) CD4+, IFNγ+ CD4+ and IL-17+CD4+ T cells in colons of germ free mice after FMT from propyzamide- or vehicle-treated mice. (n = 10 mice for vehicle CD4+ and IFNγ+ CD4+, n = 9 for vehicle IL-17+CD4+, n = 8 mice for all propyzamide groups). Data shown as mean±SEM. ***p < 0.001, ** p < 0.01, *p < 0.05.
Extended Data Fig. 4 |. Transcriptional analysis of colonic T cells and DCs.
(a) Tnf, Il23, Il1b and Il6 expression determined by qPCR in LPMC from naive mice (n = 7) and vehicle- or propyzamide-treated mice during TNBS-induced colitis (n = 6 mice per group). (b) Tgfb and Il10 expression determined by qPCR in LPMC from naive mice (n = 7) and vehicle- or propyzamide-treated mice during TNBS-induced colitis (n = 6 mice per group). Data shown as mean±SEM. (c) Dot plot visualization of features that define cell clusters in Fig. 2i. (d) UMAP plots of colonic cells from naive or TNBS-induced colitis mice treated with vehicle or propyzamide (100 mg kg−1). (e) Cluster distribution per replicates of colonic cells from naive or TNBS-induced colitis mice treated with vehicle or propyzamide (n = 5 mice per group). (f) Heatmap of differentially expressed genes that cluster colonic DC populations from scRNAseq analysis. (g) UMAP plots of DCs from colons from naive or TNBS-induced colitis mice treated with vehicle or propyzamide (100 mg kg−1). (h) GSEA analysis showing pathways activated in DCs from propyzamide-treated mice during TNBS-colitis. (i) Percentage of each DC subpopulation from vehicle- or propyzamide-treated naive mice. (j) mRNA expression determined by bulk RNA-seq in colon samples from vehicle- or propyzamide-treated mice 24 h after anti-CD3 administration (n = 4 mice per group). (k) Cyp1a1 and Cyp1b1 expression in colonic CD45+ cells from vehicle- or propyzamide-treated mice 24 h after anti-CD3 injection (n = 6 mice per group). (l) IPA showing pathways significantly upregulated in propyzamide-treated mice analysed by bulk-RNA-seq. (m,n) Rela and Cebpb (m) and Ifng, Il17, Rorc and Il12rb1 (n) expression in colonic CD45+ cells from vehicle- and propyzamide-treated mice 24 h after anti-CD3 administration (n = 10 mice for Cebpb vehicle, n = 8 mice for Cebpb propyzamide, n = 5 for Ifng vehicle, n = 4 for Ifng propyzamide, n = 5 for Il17 propyzamide, n = 5 for Rorc vehicle, n = 5 for Il12rb1 vehicle, n = 6 mice otherwise). (o) T cells, IFNγ+ and IL-17+ CD4 T cells and IFNγ+ CD8 T cells in colon from propyzamide- and vehicle-treated mice 24 h after anti-CD3 injection. Data shown as mean±SEM. ***p < 0.001, ** p < 0.01, *p < 0.05.
Extended Data Fig. 5 |. Effects of propyzamide on the small intestine.
(a) mRNA expression determined by bulk RNA-seq in jejunal CD45+ cells from vehicle- and propyzamide-treated mice (n = 4 mice per group. (b,c,d,e) CD4, IFNγ+ CD4 and IL-17+ CD4 T cells (b), CD8 and IFNγ+ CD8 T cells (c), γδ and IL-17+ γδ T cells (d), ILC and IL-17+ILC3 (e) in jejunum from vehicle- or propyzamide-treated mice. (n = 10 mice for IL-17+CD4 T cells vehicle, IFNγ+ CD8 T cells vehicle, γδ and IL-17+ γδ T cells vehicle, ILC and IL-17+ILC3 vehicle, n = 7 mice for CD8 T cells propyzamide and γδ T cells propyzamide, n = 8 for ILC propyzamide, n = 9 mice otherwise). (f) Transactivation of the RARα promoter in RARa-luciferase transfected HEK293T cells treated with retinoic acid or retinoic acid and propyzamide. (g) Transactivation of the PPARα promoter in PPARa-luciferase transfected HEK293T cells treated with fenofibrate, or propyzamide and fenofibrate for 24 h. (h) Rorc and Il12rb1 expression evaluated by qPCR in colonic CD4 T cells sorted from vehicle- or propyzamide-treated WT or AHRd mice after TNBS-colitis. (i) Il1b, Tnf and Il23 expression in sorted DCs. (j) Rela expression in BMDCs (n = 5 per group). (k) C/EBPβ expression, determined by ELISA, following Rela knockdown in BMDCs (n = 3 per group). (l) Relative expression of p65 subunit of NF-κB in primary murine DCs as a result of the depicted chemical treatment previously identified to be linked to IBD in Fig. 1d. (m) Microtubule destabilization after paclitaxel and/or propyzamide incubation with fluorescent tubulin. Data shown as mean±SEM. ****p < 0.0001, ***p < 0.001, ** p < 0.01, *p < 0.05.
Extended Data Fig. 6 |. C/EBPβ activation in DCs boosts colitogenic T-cell differentiation.
(a) Cebpb expression following propyzamide treatment in primary DCs from WT and Cebpb−/− mice (n = 14 vehicle-treated WT cells, n = 9 propyzamide-treated WT cells, n = 4 vehicle-treated Cebpb−/− cells, n = 5 propyzamide-treated Cebpb−/− cells). One-way ANOVA test followed by Tukey’s post-hoc test. (b) Il1b, Il23 and Tnf, expression following Cebpb knockdown in BMDCs. (For Il1b n = 6 siNT vehicle, n = 4 siNT propyzamide, n = 5 siCebpb vehicle and n = 6 siCebpb propyzamide. For Tnf, n = 6 siNT vehicle, n = 3 siNT propyzamide, n = 6 siCebpb vehicle and n = 6 siCebpb propyzamide. For Il23, n = 5 siNT vehicle, n = 3 siNT propyzamide, n = 7 siCebpb vehicle and n = 9 siCebpb propyzamide). One-way ANOVA test followed by Holm-Šidák’s post-hoc test. (c) Cebpb expression following Cebpb silencing in BMDCs (n = 8 siNT vehicle, n = 5 siNT propyzamide, n = 9 siCebpb vehicle and n = 5 siCebpb propyzamide). (d) Gate strategy used to sort human DCs from PBMCs. (e) CEBPB expression in human DCs treated with propyzamide after knockdown with CEBPB-targeting (siCEBPB) or non-targeting (siNT) siRNAs (n = 6 per group). Two-way ANOVA followed by Tukey’s multiple comparisons post-hoc test. (f) ChIP-seq and ATAC-seq re-analyses from23 of Cebpb binding to Il1b, Tnf and Il23 promoter in GM-CSF or Flt3 differentiated DCs. (g) Analysis of splenic DCs from TNBS-induced colitis mice. This schematic was created using BioRender. (h) IFNγ, IL-17 and TNF representative histograms in T cells co-cultured with splenic DCs from vehicle- or propyzamide-treated TNBS-induced colitis mice. MFI of IL-17, IFNγ and TNF OT-II CD4+ T cells following co-culture with splenic DCs from propyzamide- or vehicle-treated mice (n = 6 vehicle-treated mice, n = 4 propyzamide-treated mice). Unpaired t-test. (i) Ifng and Il17 expression determined by qPCR in co-cultures from WT and Cebpb−/− DCs pre-treated with vehicle or propyzamide in presence of OVA peptide and co-culture with OT-II naive CD4 T cells for 48 h (For Ifng, n = 4 WT vehicle, n = 4 WT propyzamide, n = 7 Cebpb−/− vehicle and n = 6 Cebpb−/− propyzamide. For Il17, n = 7 WT vehicle, n = 4 WT propyzamide, n = 4 Cebpb−/− vehicle and n = 6 Cebpb−/− propyzamide). One-way ANOVA test followed by Holm-Šidák’s multiple comparisons post-hoc test. (j) TNF, IL-17 and IL-6 relative levels determined in co-culture supernatants of human DCs pre-treated as specified in the figure, and then incubated with allogenic T cells for 48 h. (For Tnf, n = 3 per group. For Il17, n = 4 per group). One-way ANOVA test followed by Holm-Šidák’s multiple comparisons post-hoc test. (k–l) Cebpb expression (k) and Il1b, Il23 and Tnf expression (l) determined by qPCR in primary splenic DCs transfected with cumate-inducible Cebpb-expressing plasmid after cumate treatment as depicted for 96 h. (For Cebpb (k), n = 5 vehicle-treated and n = 3 for each cumate-treated group. For Tnf and Il23 (l) n = 4 vehicle-treated and n = 3 for each cumate-treated group). One-way ANOVA test followed by Holm-Šidák’s post-hoc test. Data shown as mean±SEM.
Extended Data Fig. 7 |. scRNAseq analysis of intestinal DCs from IBD patients.
(a) UMAP plot and cluster distribution per replicates of colonic leukocytes isolated from Cebpb−/− (n = 2 mice) and WT (n = 5 mice) DC chimeras after TNBS-colitis. (b) UMAP plot showing DC clusters analysed in colon from WT and Cebpb−/− DC chimeras analysed by scRNAseq. (c) Number of cells per DC cluster and cluster distribution per replicates. (d) Cebpb expression in DCs recovered from WT or Cebpb−/− DC chimera. (e) UMAP plot of 58 samples from 44 IBD patients and healthy controls. (f) Dot Plot visualization of features that define DCs and T cells. (g) Differentially regulated pathways in DCs from IBD patients. (h) Violin plot depicting CEBPB expression in DCs from IBD and healthy controls. (i) UMAP depicting intestinal DCs from IBD and healthy controls analysed by scRNA-seq. (j) CEBPB expression in DC1 and DC2 subsets. (k) Differentially regulated pathways in CEBPB expressing DCs.
Extended Data Fig. 8 |. VCAM-1 blockade ameliorates intestinal inflammation.
(a) Network analysis of molecules reported in ToxCast database to be induced by propyzamide. (b) Effect of propyzamide on Vcam1 promoter activity (n = 10 for 0.2 μM propyzamide, n = 7 otherwise). (c) Vcam1 expression in colons from naive and TNBS-induced colitis mice treated with vehicle or propyzamide (100 mg kg−1) (n = 5 mice per group). Unpaired t-test. (d) Effect of propyzamide on Itga4 and Itga1 expression in T cells (n = 3 for Itga4 propyzamide, n = 4 otherwise). Unpaired t test. (e) Evaluation of VCAM-1 blocking antibodies in TNBS-induced colitis. This schematic was created using BioRender. (f) Effect of VCAM-1 blocking antibodies (100 μg per mouse) or isotype controls on TNBS-induced colitis (Weight: n = 12 for vehicle isotype control, n = 9 for vehicle anti-VCAM1, n = 7 for propyzamide anti-VCAM1, n = 13 otherwise, Colon length: n = 15 for naive, n = 5 for vehicle isotype control, n = 13 for propyzamide isotype control, n = 7 otherwise). (g,h) Representative hematoxylin & eosin staining (g) and clinical histomorphology scores (h) (n = 4 mice for antiVCAM1-treated mice, n = 5 mice otherwise). Arrows show leukocyte infiltrates. (i) Effect of VCAM-1 blocking antibodies (100 μg per mouse) or isotype controls on colonic IFNγ+ and IL17+ CD4+ T cells during TNBS-induced colitis determined by flow cytometry (IFNγ: n = 6 mice for vehicle isotype control, n = 5 mice for propyzamide isotype control, n = 4 otherwise. IL17: n = 4 mice for vehicle and propyzamide anti-VCAM1, n = 5 otherwise). One-way ANOVA followed by post-hoc tests Tukey’s or Holm-Sidak’s test for selected multiple comparisons for b, f, h and i. Data shown as mean±SEM.
Extended Data Fig. 9 |. Propyzamide boosts colitogenic T-cell differentiation.
(a) Gate strategy used to study CD4 T cells. (b) Percentage of IFNγ+ CD4+ T cells and Ifng expression of CD4+ T cells activated under Th1-polarizing conditions in the presence of vehicle or propyzamide (n = 3 per group). Unpaired t-test. (c) Percentage of IL-17+ CD4+ T cells and Il17 expression of CD4+ T cells activated under Th17-polarizing conditions in the presence of vehicle or propyzamide (n = 3 per group). Unpaired t-test. (d) Tbx21, Csf2 and Il12rb1 expression in CD4+ T cells activated under Th1 polarizing conditions in the presence of vehicle or propyzamide (n = 3 per group, except propyzamide-treated cells expressing Il12rb1, n = 4). Unpaired t-test (e) Csf2, Tnf and Rorc expression in CD4+ T cells activated under Th17 polarizing conditions in the presence of vehicle or propyzamide (For Csf2 n = 3per group, for Rorc n = 3 vehicle-treated and n = 4 propyzamide-treated, for Tnf n = 5 per group) Unpaired t-test. (f) Nuclear p65 translocation in splenic CD4+ T cells activated with anti-CD3 and anti-CD28 in the presence of vehicle or propyzamide (n = 3 per group). Unpaired t-test. (g,h) Rela (g) and Cebpb (h) expression following Rela knockdown in T cells and treated with vehicle or propyzamide (For (g) n = 3 per siNT group and n = 4 per siRela group, for (h) n = 5 per siNT group and n = 4 per siRela group). One-way ANOVA test followed by Holm-Šidák’s multiple comparisons post-hoc test. (i) Cebpb expression in murine splenic WT and Cebpb−/− T cells (n = 9 vehicle-treated WT cells, n = 7 propyzamide-treated WT cells, n = 3 vehicle-treated Cebpb−/− cells, n = 3 propyzamide-treated Cebpb−/− cells). One-way ANOVA test followed by Holm-Šidák’s multiple comparisons post-hoc test. (j) CEBPB expression following CEBPB knockdown in human T cells.(n = 4 per group except n = 3 siCEBPB propyzamide-treated group). (k) ILCs and CD8 T cells in colons from Rag1−/− mice reconstituted with WT or Cebpb−/− T cells. (n = 4 per group except Cebpb−/− CD8 T cells n = 5). Data shown as mean±SEM. ***p < 0.001, ** p < 0.01, *p < 0.05.
Extended Data Fig. 10 |. scRNAseq analysis of intestinal T cells from IBD patients.
(a) UMAP depicting total intestinal T cells analysed by scRNA-seq. (b) Dot plot visualization of features to identify T-cell subsets. (c) Heat map of differentially expressed genes in T cells from IBD patients and healthy controls. (d) Upstream analysis of NF-κB-driven CEBPB expression in T cells. (e) CEBPB expressing T cells from IBD and HC samples. (f,g,h) Pathway analysis of differentially expressed genes in Resident Memory T cells (f), CD8 T cells (g) and ILCs (h) from IBD and healthy control samples. (i) Graphical model of modulation of colitogenic T cell responses by NF-κB-driven C/EBPβ signalling and propyzamide.
Supplementary Material
Acknowledgements
We thank all of the members of the Quintana laboratory for advice and discussions; R. Krishnan for technical assistance with flow cytometry studies; E. Buys and the BWH aquatics facility for assistance with breeding and maintaining the zebrafish; the staff at the Tufts and Harvard histology core facilities for providing histopathology services; the staff at the NeuroTechnology Studio at Brigham and Women’s Hospital for providing instrument access; L. Zon and G. Stirtz at Boston Children’s Hospital for providing zebrafish lines and advice; D. Rojas Marquez (@darwid_illustration) for help with the model figure. All of the other illustrations were created using BioRender. This work was supported by grants NS087867, ES025530, ES032323, AI126880 and AI093903 from the National Institutes of Health. C.-C.C. received support (104-2917-I-564 -024) from the Ministry of Science and Technology, Taiwan. Y.-C.W. received support by grants and 109-2221-E-010-013-MY3 and 107-2221-E-010-019-MY3 from the Ministry of Science and Technology, Taiwan. C.M.P. was supported by a fellowship from the FAPESP BEPE (2019/13731-0). C.G.-V. was supported by an Alfonso Martín Escudero Foundation postdoctoral fellowship and by a postdoctoral fellowship from the European Molecular Biology Organization (ALTF 610-2017). G.P. is a trainee in the Medical Scientist Training Program funded by NIH T32 GM007356. The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Science or NIH. H.-G.L. was supported by a Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2021R1A6A3A14039088). M.J. was supported by a post-doctoral fellowship from Sigrid Juselius, personal post-doctoral grants from Saastamoinen Foundation, Paulo Foundation, The Finnish MS-Foundation, Orion Farmos Research Foundation and Maud Kuistila Memory Foundation. M.A.W. was supported by the NIH (1K99NS114111, F32NS101790), a training grant from the NIH and Dana-Farber Cancer Institute (T32CA207201), a travelling neuroscience fellowship from the Program in Interdisciplinary Neuroscience at the Brigham and Women’s Hospital and the Women’s Brain Initiative at the Brigham and Women’s Hospital. V.R. received support from an educational grant from Mallinkrodt Pharmaceuticals (A219074) and by a fellowship from the German Research Foundation (DFG RO4866 1/1). R.C. received support by a postdoctoral fellowship from the Swedish Research Council. B.M.A. received support from K12CA090354 from the NIH.
Footnotes
Competing interests The authors declare no competing interests.
Additional information
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41586-022-05308-6.
Peer review information Nature thanks Judy Cho, Mark Sundrud and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Reprints and permissions information is available at http://www.nature.com/reprints.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41586-022-05308-6.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
RNA-seq and scRNA-seq data have been deposited at the GEO database under the following accession number GSE194412 and GSE175766. 16S rRNA-sequencing data have been submitted to the NCBI sequence-read archive under BioProject number PRJNA804134. The machine learning codes used for this study can be accessed at https://github.com/Quin-tanaLab/IBD_function_public. Source data are provided with this paper.
References
- 1.Huang H et al. Fine-mapping inflammatory bowel disease loci to single-variant resolution. Nature 547, 173–178 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kamm MA Rapid changes in epidemiology of inflammatory bowel disease. Lancet 390, 2741–2742 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Covacu R et al. System-wide analysis of the T cell response. Cell Rep. 14, 2733–2744 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quintana FJ et al. Adaptive autoimmunity and Foxp3-based immunoregulation in zebrafish. PLoS ONE 5, e9478 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wheeler MA et al. Environmental control of astrocyte pathogenic activities in CNS inflammation. Cell 176, 581–596 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.White RM et al. DHODH modulates transcriptional elongation in the neural crest and melanoma. Nature 471, 518–522 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scott BM et al. Self-tunable engineered yeast probiotics for the treatment of inflammatory bowel disease. Nat. Med. 27, 1212–1222 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Fleming A, Jankowski J & Goldsmith P In vivo analysis of gut function and disease changes in a zebrafish larvae model of inflammatory bowel disease: a feasibility study. Inflamm. Bowel Dis. 16, 1162–1172 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Goettel JA et al. AHR activation is protective against colitis driven by T cells in humanized mice. Cell Rep. 17, 1318–1329 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richard AM et al. ToxCast chemical landscape: paving the road to 21st century toxicology. Chem. Res. Toxicol. 29, 1225–1251 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Gut P et al. Whole-organism screening for gluconeogenesis identifies activators of fasting metabolism. Nat. Chem. Biol. 9, 97–104 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.North TE et al. Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature 447, 1007–1011 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richter S, Schulze U, Tomancak P & Oates AC Small molecule screen in embryonic zebrafish using modular variations to target segmentation. Nat. Commun. 8, 1901 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keiser MJ et al. Predicting new molecular targets for known drugs. Nature 462, 175–181 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chassaing B et al. Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature 519, 92–96 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaakoush NO Sutterella species, IgA-degrading bacteria in ulcerative colitis. Trends Microbiol. 28, 519–522 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Sanmarco LM et al. Gut-licensed IFNγ+ NK cells drive LAMP1+TRAIL+ anti-inflammatory astrocytes. Nature 590, 473–479 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schiering C et al. Feedback control of AHR signalling regulates intestinal immunity. Nature 542, 242–245 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rothhammer V et al. Microglial control of astrocytes in response to microbial metabolites. Nature 557, 724–728 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okey AB, Vella LM & Harper PA Detection and characterization of a low affinity form of cytosolic Ah receptor in livers of mice nonresponsive to induction of cytochrome P1–450 by 3-methylcholanthrene. Mol. Pharmacol. 35, 823–830 (1989). [PubMed] [Google Scholar]
- 21.Akashi TI, Nagano K, Enomoto E, Mizuno M & Shibaok K Effects of propyzamide on tobacco cell microtubules in vivo and in vitro. Plant Cell Physiol. 29, 1053–1062 (1988). [Google Scholar]
- 22.Jackman RW, Rhoads MG, Cornwell E & Kandarian SC Microtubule-mediated NF-κB activation in the TNF-α signaling pathway. Exp. Cell. Res. 315, 3242–3249 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garber M et al. A high-throughput chromatin immunoprecipitation approach reveals principles of dynamic gene regulation in mammals. Mol. Cell 47, 810–822 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Satpathy AT et al. Notch2-dependent classical dendritic cells orchestrate intestinal immunity to attaching-and-effacing bacterial pathogens. Nat. Immunol. 14, 937–948 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meredith MM et al. Expression of the zinc finger transcription factor zDC (Zbtb46, Btbd4) defines the classical dendritic cell lineage. J. Exp. Med. 209, 1153–1165 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elmentaite R et al. Single-cell sequencing of developing human gut reveals transcriptional links to childhood Crohn’s disease. Dev. Cell 55, 771–783 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin JC et al. Single-cell analysis of Crohn’s disease lesions identifies a pathogenic cellular module associated with resistance to anti-TNF therapy. Cell 178, 1493–1508 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boland BS et al. Heterogeneity and clonal relationships of adaptive immune cells in ulcerative colitis revealed by single-cell analyses. Sci. Immunol. 5, eabb4432 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cybulsky MI et al. Gene structure, chromosomal location, and basis for alternative mRNA splicing of the human VCAM1 gene. Proc. Natl Acad. Sci. USA 88, 7859–7863 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oh H & Ghosh S NF-κB: roles and regulation in different CD4+ T-cell subsets. Immunol. Rev. 252, 41–51 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chu H et al. Gene-microbiota interactions contribute to the pathogenesis of inflammatory bowel disease. Science 352, 1116–1120 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lamas B et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat. Med. 22, 598–605 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rothschild D et al. Environment dominates over host genetics in shaping human gut microbiota. Nature 555, 210–215 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Iyer SS et al. Dietary and microbial oxazoles induce intestinal inflammation by modulating aryl hydrocarbon receptor responses. Cell 173, 1123–1134 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cole DJ Metabolic Pathways of Agrochemicals. Part One—Herbicides and Plant Growth Regulators (eds Roberts T et al.) (Royal Society of Chemistry, 1998). [Google Scholar]
- 36.Propyzamide; Pesticide Tolerances; https://www.federalregister.gov/documents/2016/01/13/2016-00534/propyzamide-pesticide-tolerances (US Government, 2016). [Google Scholar]
- 37.Chaiklieng S, Suggaravetsiri P & Autrup H Risk assessment on benzene exposure among gasoline station workers. Int. J. Environ. Res. Publ. Health 16, 2545 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ott MG, Diller WF & Jolly AT Respiratory effects of toluene diisocyanate in the workplace: a discussion of exposure-response relationships. Crit. Rev. Toxicol. 33, 1–59 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Cuenca L et al. Environmentally-relevant exposure to diethylhexyl phthalate (DEHP) alters regulation of double-strand break formation and crossover designation leading to germline dysfunction in Caenorhabditis elegans. PLoS Genet. 16, e1008529 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.World Health Organization. Guidelines for Drinking-Water Quality Vol. 2, Ch. 14.11, 461–467 (1996). [Google Scholar]
- 41.Toxicological Profile for Toluene Diisocyanate and Methylenediphenyl Diisocyanate (US Department of Health and Human Services, 2018). [PubMed] [Google Scholar]
- 42.World Health Organization. Guidelines for Drinking-Water Quality Vol. 2, Ch. 14.21, 530–540 (1996). [Google Scholar]
- 43.Sorg O AhR signalling and dioxin toxicity. Toxicol. Lett. 230, 225–233 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Muku GE, Murray IA, Espín JC & Perdew GH Urolithin A is a dietary microbiota-derived human aryl hydrocarbon receptor antagonist. Metabolites 8, 86 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gerondakis S, Fulford TS, Messina NL & Grumont RJ NF-κB control of T cell development. Nat. Immunol. 15, 15–25 (2014). [DOI] [PubMed] [Google Scholar]
- 46.Balasubramani A et al. Modular utilization of distal cis-regulatory elements controls Ifng gene expression in T cells activated by distinct stimuli. Immunity 33, 35–47 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ruan Q et al. The Th17 immune response is controlled by the Rel-RORγ-RORγ T transcriptional axis. J. Exp. Med. 208, 2321–2333 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yosef N et al. Dynamic regulatory network controlling TH17 cell differentiation. Nature 496, 461–468 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jostins L et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491, 119–124 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Satoh T et al. Identification of an atypical monocyte and committed progenitor involved in fibrosis. Nature 541, 96–101 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Jaronen M, Wheeler MA & Quintana FJ Protocol for inducing inflammation and acute myelin degeneration in larval zebrafish. STAR Protoc. 3, 101134 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nüsslein-Volhard C & Dahm R Zebrafish: A Practical Approach 1st edn (Oxford Univ. Press, 2002). [Google Scholar]
- 53.Cusick MF, Libbey JE, Trede NS, Eckels DD & Fujinami RS Human T cell expansion and experimental autoimmune encephalomyelitis inhibited by Lenaldekar, a small molecule discovered in a zebrafish screen. J. Neuroimmunol. 244, 35–44 (2012). [DOI] [PubMed] [Google Scholar]
- 54.Ridges S et al. Zebrafish screen identifies novel compound with selective toxicity against leukemia. Blood 119, 5621–5631 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.ToxCast & Tox21 Summary Files from invitrodb_v3; https://www.epa.gov/chemical-research/toxicity-forecaster-toxcasttm-data (US EPA, accessed 28 October 2018). [Google Scholar]
- 56.Ruder B, Atreya R & Becker C Tumour necrosis factor alpha in intestinal homeostasis and gut related diseases. Int. J. Mol. Sci. 20, 1887 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Andreou NP, Legaki E & Gazouli M Inflammatory bowel disease pathobiology: the role of the interferon signature. Ann. Gastroenterol. 33, 125–133 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McEntee CP, Finlay CM & Lavelle EC Divergent roles for the IL-1 family in gastrointestinal homeostasis and inflammation. Front. Immunol. 10, 1266 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Salas A et al. JAK-STAT pathway targeting for the treatment of inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 17, 323–337 (2020). [DOI] [PubMed] [Google Scholar]
- 60.Decara J et al. Peroxisome proliferator-activated receptors: experimental targeting for the treatment of inflammatory bowel diseases. Front. Pharmacol. 11, 730 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pernomian L, Duarte-Silva M & de Barros Cardoso CR The aryl hydrocarbon receptor (AHR) as a potential target for the control of intestinal inflammation: insights from an immune and bacteria sensor receptor. Clin. Rev. Allergy Immunol. 59, 382–390 (2020). [DOI] [PubMed] [Google Scholar]
- 62.Langfelder P & Horvath S Eigengene networks for studying the relationships between co-expression modules. BMC Syst. Biol. 1, 54 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Martínez-Camblor P, Pérez-Fernández S & Díaz-Coto S The role of the p-value in the multitesting problem. J. Appl. Stat. 47, 1529–1542 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Breiman L Random forests. Mach. Learn. 45, 5–32 (2001). [Google Scholar]
- 65.Tong H, Faloutsos C & Pan J Fast random walk with restart and its applications. In Proc. Sixth International Conference on Data Mining (ICDM’06) 613–622 (IEEE, 2006). [Google Scholar]
- 66.Kohler S, Bauer S, Horn D & Robinson PN Walking the interactome for prioritization of candidate disease genes. Am. J. Hum. Genet. 82, 949–958 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang B & Horvath S A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 4, 17 (2005). [DOI] [PubMed] [Google Scholar]
- 68.Zhou G et al. NetworkAnalyst 3.0: a visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 47, W234–W241 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Neurath MF, Fuss I, Kelsall BL, Stuber E & Strober W Antibodies to interleukin 12 abrogate established experimental colitis in mice. J. Exp. Med. 182, 1281–1290 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Soumillon M, Cacchiarelli D, Semrau S, van Oudenaarden A & Mikkelsen TS Characterization of directed differentiation by high-throughput single-cell RNA-Seq. Preprint at bioRxiv 10.1101/003236 (2014). [DOI] [Google Scholar]
- 71.Dobin A et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li B & Dewey CN RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bray NL, Pimentel H, Melsted P & Pachter L Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527 (2016). [DOI] [PubMed] [Google Scholar]
- 74.Love MS, Soneson C & Robinson MD Importing transcript abundance datasets with tximport. Bioconductor https://bioconductor.org/packages/devel/bioc/vignettes/tximport/inst/doc/tximport.html (2017). [Google Scholar]
- 75.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhu A, Ibrahim JG & Love MI Heavy-tailed prior distributions for sequence count data: removing the noise and preserving large differences. Bioinformatics 35, 2084–2092 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zheng GX et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wolock SL, Lopez R & Klein AM Scrublet: computational identification of cell doublets in single-cell transcriptomic data. Cell Syst. 8, 281–291 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hafemeister C & Satija R Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20, 296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Butler A, Hoffman P, Smibert P, Papalexi E & Satija R Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Korsunsky I et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mi H, Muruganujan A, Ebert D, Huang X & Thomas PD PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 47, D419–D426 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Finak G et al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 16, 278 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Caporaso JG et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Walters W et al. Improved bacterial 16S rRNA gene (V4 and V4–5) and fungal internal transcribed spacer marker gene primers for microbial community surveys. mSystems 1, e00009–15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cox LM et al. Calorie restriction slows age-related microbiota changes in an Alzheimer’s disease model in female mice. Sci. Rep. 9, 17904 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Caporaso JG et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yoon SH et al. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 67, 1613–1617 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lozupone C, Lladser ME, Knights D, Stombaugh J & Knight R UniFrac: an effective distance metric for microbial community comparison. ISME J. 5, 169–172 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chu C et al. The microbiota regulate neuronal function and fear extinction learning. Nature 574, 543–548 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yeste A et al. Tolerogenic nanoparticles inhibit T cell-mediated autoimmunity through SOCS2. Sci. Signal. 9, ra61 (2016). [DOI] [PubMed] [Google Scholar]
- 92.Rothhammer V et al. Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat. Med. 22, 586–597 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Burbach KM, Poland A & Bradfield CA Cloning of the Ah receptor cDNA reveals a distinctive ligand-activated transcription factor. Proc. Natl Acad. Sci. USA 89, 8185–8189 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dolwick KM, Schmidt JV, Carver LA, Swanson HI & Bradfield CA Cloning and expression of a human Ah receptor cDNA. Mol. Pharmacol. 44, 911–917 (1993). [PubMed] [Google Scholar]
- 95.Lowe MM et al. Identification of cinnabarinic acid as a novel endogenous aryl hydrocarbon receptor ligand that drives IL-22 production. PLoS ONE 9, e87877 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Song J et al. A ligand for the aryl hydrocarbon receptor isolated from lung. Proc. Natl Acad. Sci. USA 99, 14694–14699 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Parks AJ et al. In silico identification of an aryl hydrocarbon receptor (AHR) antagonist with biological activity in vitro and in vivo. Mol. Pharmacol. 86, 593–608 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mascanfroni ID et al. IL-27 acts on DCs to suppress the T cell response and autoimmunity by inducing expression of the immunoregulatory molecule CD39. Nat. Immunol. 14, 1054–1063 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Joung J et al. Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat. Protoc. 12, 828–863 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq and scRNA-seq data have been deposited at the GEO database under the following accession number GSE194412 and GSE175766. 16S rRNA-sequencing data have been submitted to the NCBI sequence-read archive under BioProject number PRJNA804134. The machine learning codes used for this study can be accessed at https://github.com/Quin-tanaLab/IBD_function_public. Source data are provided with this paper.