Abstract
Teleost fishes are ancient tetraploids descended from an ancestral whole-genome duplication that may have contributed to the impressive diversification of this clade. Whole-genome duplications can occur via self-doubling (autopolyploidy) or via hybridization between different species (allopolyploidy). The mode of tetraploidization conditions evolutionary processes by which duplicated genomes return to diploid meiotic pairing, and subsequent genetic divergence of duplicated genes (cytological and genetic rediploidization). How teleosts became tetraploid remains unresolved, leaving a fundamental gap in the interpretation of their functional evolution. As a result of the whole-genome duplication, identifying orthologous and paralogous genomic regions across teleosts is challenging, hindering genome-wide investigations into their polyploid history. Here, we combine tailored gene phylogeny methodology together with a state-of-the-art ancestral karyotype reconstruction to establish the first high-resolution comparative atlas of paleopolyploid regions across 74 teleost genomes. We then leverage this atlas to investigate how rediploidization occurred in teleosts at the genome-wide level. We uncover that some duplicated regions maintained tetraploidy for more than 60 million years, with three chromosome pairs diverging genetically only after the separation of major teleost families. This evidence suggests that the teleost ancestor was an autopolyploid. Further, we find evidence for biased gene retention along several duplicated chromosomes, contradicting current paradigms that asymmetrical evolution is specific to allopolyploids. Altogether, our results offer novel insights into genome evolutionary dynamics following ancient polyploidizations in vertebrates.
Since the first teleost fish genome sequence was published in 2002 (Aparicio et al. 2002), fish genomics has massively contributed to our understanding of vertebrate genome function and evolution. As an early-diverging vertebrate clade, teleosts are at an ideal phylogenetic position to conduct comparative analyses with tetrapods and study deep-rooting vertebrate features. The conservation of regulatory circuits and developmental pathways has turned zebrafish, medaka, and—to a lesser extent—platyfish into model species for human diseases (Wittbrodt et al. 2002; Lieschke and Currie 2007; Schartl 2014). In addition, fish have become popular in evolutionary, ecological, and physiological genomics, illuminating processes such as environmental adaptation, species diversification, social behavior, or sex determination (Rittschof et al. 2014; Capel 2017; Salzburger 2018; Kim et al. 2019; Xie et al. 2019; Greenway et al. 2020). Around two hundred fish species have reference genome assemblies, and many more are expected to become available in the coming years (Rhie et al. 2021), requiring improved comparative frameworks to dissect the functional evolution of teleost genomes.
All teleost fish species are paleopolyploids descended from an ancient round of whole-genome duplication (WGD), dated at approximately 320 Mya (Jaillon et al. 2004). This evolutionary event, referred to as the teleost-specific genome duplication (TGD), doubled all chromosomes and genes present in the teleost ancestor. The TGD has left a significant imprint on extant teleost genomes: although most duplicated genes have returned to a single-copy state, a high fraction of teleost genes remain in two copies, called ohnologs. For instance, 26% of all zebrafish genes are still retained as duplicated ohnologs (Howe et al. 2013). Evidence suggests that TGD duplicates have been involved in the evolution of innovations (Zakon et al. 2006; Moriyama et al. 2016; Escobar-Camacho et al. 2020), but it remains unclear how differential gene retention and functional divergence have sustained the impressive phenotypic diversity of the teleost clade.
WGD can arise through two mechanisms: autopolyploidization (within or between populations of a single species) or allopolyploidization (after hybridization of related species), with different consequences on subsequent genome evolution (Stebbins 1947; Mason and Wendel 2020). In particular, auto- and allopolyploidization differentially shape the rediploidization process, that is, how the polyploid genome returns to a largely diploid state over time. Because of the initial sequence similarity between duplicated chromosomes (homeologs), young autopolyploid genomes are characterized by multivalent meiotic behavior and tetrasomic inheritance, with recombination and gene conversion occurring between homeologous chromosomes. Cytological rediploidization, that is, the return to bivalent meiotic pairing, is necessary to enable the diploid inheritance of two distinct duplicated regions in the genome. The mechanisms driving cytological rediploidization remain poorly understood, with proposed roles for genomic rearrangements and reduced crossing-over frequency (Bomblies et al. 2016; Mandáková and Lysak 2018). Maintenance of tetraploidy for millions of years over entire homeologs has been previously shown following autotetraploidization events in salmonids and Acipenseriformes (Robertson et al. 2017; Gundappa et al. 2022; Redmond et al. 2022). In these clades, rediploidization has been an extended process, with some duplicated genomic regions quickly restoring diploid behavior, whereas others have maintained tetraploidy for tens of millions of years. In contrast, after allopolyploidization events, the extent of duplicated chromosome pairing depends on genetic similarity between the parental genomes. Localized homeologous exchanges, in which sequences from one subgenome are substituted by sequences from the other, have been observed in allopolyploids, including allopolyploid crops and paleopolyploid carps (Lloyd et al. 2018; Li et al. 2021). These events typically concern a minor fraction of allopolyploid genomes (<3%), and have never been shown to be maintained over millions of years.
The retention and divergence of gene copies is also affected by the nature of the polyploidization event. After allotetraploidization, one of the two subgenomes often loses more genes than the other (Garsmeur et al. 2014; Session et al. 2016; Cheng et al. 2018), although not always (Sun et al. 2017). This unequal gene retention has been linked to differences in transposable element repertoires and epigenetic silencing in the two subgenomes, and orients further functional evolution. Such differences can lead to reduced expression levels and relaxed selective pressure biased toward one of the subgenomes, which then accumulates more gene losses (Freeling et al. 2012; Bird et al. 2021). Conversely, in the case of autopolyploidy, gene losses are expected to affect both homeologs equally owing to their high similarity. It remains unclear whether the teleost whole-genome duplication corresponds to an ancestral auto- or an allotetraploidization event, a significant gap in our understanding of the early vertebrate evolutionary processes that led to the diversification of teleosts (Martin and Holland 2014; Conant 2020).
The redundancy in fish genomes can be appreciated at the macrosyntenic level, in which remnants of ancestrally duplicated chromosomes form runs of large duplicated regions known as double-conserved syntenic (DCS) regions (Postlethwait et al. 2000; Taylor et al. 2003; Jaillon et al. 2004). The precise identification and delimitation of teleost DCS regions is the key to reconstructing how rediploidization occurred and its associated impact on teleost evolution. However, WGDs present severe challenges to both gene phylogeny and ancestral genome reconstruction methodologies (Nakatani and McLysaght 2017; Zwaenepoel and Van de Peer 2019; Parey et al. 2020). Previous characterizations of DCS regions in teleosts were therefore limited to regions of highly conserved gene order or small species sets: the largest multispecies data set comprised eight fish species and included ∼29% of all genes in DCS regions (Conant 2020).
Here, we apply a phylogenetic pipeline specifically developed for WGD events (Parey et al. 2020) to retrace the evolutionary history of the genes and chromosomes of 74 teleost species encompassing most of the major fish clades. We combine these phylogenetic trees with the latest ancestral reconstruction of the pre-TGD ancestral karyotype (Nakatani and McLysaght 2017) to build a comprehensive comparative atlas of TGD-duplicated regions in teleost fish genomes. We then leverage this comparative atlas of teleost genomes to reconstruct how rediploidization occurred genome-wide following the teleost genome duplication, revealing how the ancestral teleost became tetraploid.
Results
Construction of TGD-aware teleost gene trees
We collected a data set of 101 vertebrate genomes, including 74 teleost fish, two nonteleost fish (bowfin and spotted gar, which did not undergo the teleost genome duplication or TGD), 20 other vertebrates of which six are mammals, and five nonvertebrate genomes (Supplemental Fig. S1; Supplemental Table S1). We used TreeBeST to reconstruct the phylogenetic relationships of 26,692 gene families across those 101 genomes (Methods; Vilella et al. 2009; Herrero et al. 2016). We then applied SCORPiOs to correctly place the TGD in these gene trees, using the bowfin and the spotted gar as reference outgroups (Parey et al. 2020). Briefly, SCORPiOs leverages synteny information to distinguish WGD-descended orthologs from paralogs when sequence information is inconclusive. After a WGD, orthologous genes are expected to be embedded in orthologous neighborhoods. For each individual gene tree, SCORPiOs “crowd-sources” additional information from local syntenic genes to identify errors in orthology relationships. SCORPiOs then reorganizes those gene trees to accurately position the WGD duplication node, if the synteny-consistent solution is equally supported by the sequence alignment. These 26,692 WGD-aware teleost gene trees predict that the ancestral genome of teleost fish contained 46,206 genes after the duplication event, in line with the latest estimates from the Ensembl database (49,255 ancestral teleost genes in release v100; Methods).
A high-resolution atlas of the TGD duplication across 74 teleost genomes
Teleost fish genomes are mosaics of duplicated regions, formed through rearrangements of duplicated ancestral chromosomes (Supplemental Fig. S2A). Long-standing efforts have been made to reconstruct the ancestral teleost karyotype before the whole-genome duplication (Jaillon et al. 2004; Kasahara et al. 2007; Nakatani and McLysaght 2017). According to the state-of-the-art reference, this ancestral teleost karyotype comprised 13 chromosomes (Nakatani and McLysaght 2017). Nakatani and McLysaght delineated between 353 and 690 megabase-scale genomic regions that descend from these 13 ancestral chromosomes in zebrafish, tetraodon, stickleback, and medaka.
We combined this pre-TGD ancestral karyotype with our gene trees to identify regions and genes that descend from sister duplicated chromosomes across all 74 teleosts in our data set (Methods; Supplemental Fig. S2). First, we transformed the reference segmentations of the zebrafish, tetraodon, stickleback, and medaka genomes (reference species) from 13 to 26 ancestral chromosomes after the duplication (1a, 1b, …, 13a, 13b). In each genome, we iteratively grouped regions from an ancestral chromosome into two postduplication copies by minimizing intragroup paralogs (Methods; Supplemental Fig. S2B). To assess robustness, we performed 100 groupings with random restart and found that genes were assigned to the same chromosome copy in 80% of iterations on average. We then identified orthologous ancestral chromosomes across all four species using gene orthologs, and arbitrarily named one of each pair “a” and “b” (Supplemental Fig. S2C).
Next, we propagated these ancestral chromosome annotations to the other 70 teleost genomes using gene orthology relationships (Supplemental Fig. S2D). For 1303 gene trees (7%), orthologs from the four reference species were not assigned to the same ancestral chromosome, and we resolved these inconsistencies by assigning all orthologous genes, including those of the reference species, to the most represented ancestral chromosome. This process results in a 74-species comparative genomic atlas with genes annotated to postduplication chromosomes, along with fully resolved orthology and paralogy links between all included species (Supplemental Fig. S2E).
This comparative atlas integrates 70%–90% of each genome into 24,938 postduplication gene families (Fig. 1A,B), and greatly improves upon the state-of-the-art both in terms of species and genomic coverage. The atlas reveals that teleost fishes vary substantially in their retention of duplicated gene copies (ohnologs) since the TGD, which make up 33% of the arowana genome but only 19% of the cod genome (Supplemental Table S2). In general, Osteoglossiformes, Otomorpha, and Salmoniformes species have retained more ohnologs from the TGD than other Euteleosteomorpha clades that diverged later. The atlas is available on the Genomicus database web server (see “Data availability and implementation”).
Figure 1.
Comparative atlas of TGD-duplicated regions across 74 teleosts. (A) Phylogenetic tree of the 74 teleost genomes in the comparative atlas and 27 outgroups. The color map represents the proportion of genes from each species annotated in the comparative atlas. Divergence times were extracted from TimeTree (Kumar et al. 2017). (B) Karyotype paintings using the comparative atlas. At the top, we show the inferred ancestral karyotype after the teleost whole-genome duplication (TGD). Below, karyotypes of three teleost genomes are colored by their ancestral chromosome of origin according to the comparative atlas (1a, 1b, …, 13a, 13b).
Quality checking the teleost genome comparative atlas
As a quality check, we assessed discrepancies between the pre-TGD karyotype reconstruction by Nakatani and McLysaght (2017) and our ancestral chromosome assignations. Both should be globally congruent because the ancestral karyotype provides the groundwork for the comparative atlas, but discrepancies can arise when orthologous genes are assigned to different preduplication chromosomes between the four reference species. In this case, we resolve the inconsistency by reassigning all orthologs to the most represented preduplication chromosome, which will be different from the original assignation in Nakatani and McLysaght (2017) for at least one species. We found that 9% of zebrafish genes differ in ancestral chromosome assignation between our comparative map and the pre-TGD karyotype, versus 2%–3% in medaka, tetraodon, or stickleback. This likely reflects small-scale rearrangements in zebrafish captured by our individual gene trees but missed by the macrosyntenic approach of Nakatani and McLysaght (2017). Alternatively, zebrafish genes may be more frequently placed incorrectly in our gene trees and assigned to the wrong orthology group, which may lead to erroneous ancestral chromosome reassignations. To explore this possibility, we identified gene trees that remain synteny-inconsistent after correction by SCORPiOs (i.e., whose orthology relationships are inconsistent with those of their surrounding genes), and therefore potentially contain orthology errors. Zebrafish genes reassigned to a different ancestral chromosome in our comparative map are not overrepresented in synteny-inconsistent trees (7% vs. 14% for all zebrafish genes), suggesting that their orthology relationships and chromosomal reassignations are overall well-supported by their sequences and syntenic gene neighborhoods.
Additionally, we used random noise simulations to show that the comparative atlas is robust to potential uncertainty or errors in the original ancestral genome reconstruction. The delineation of genomic regions descended from each preduplication chromosome from Nakatani and McLysaght (2017) relies on the identification of conserved synteny blocks and their breakpoints between teleost genomes and outgroups. Because breakpoint locations are challenging to determine—as also attested by lower posterior probabilities close to breakpoints in Nakatani and McLysaght (2017)—the region boundaries can vary in precision. We shifted the positions of these boundaries with increasingly large errors, mimicking situations in which up to 25% of genes change preduplication chromosome assignations in each of the reference genomes (Methods). We found that even large errors in region boundaries did not majorly affect the final atlas, with only 11% of genes changing ancestral chromosome assignations at the highest noise settings (Supplemental Fig. S3).
Finally, we report that correcting gene trees with SCORPiOs had a decisive impact on the establishment of the comparative atlas, enabling the inclusion of a significantly larger fraction of teleost genes (83% vs. 61%; Supplemental Fig. S4). The teleost comparative genomic atlas represents therefore a reliable, comprehensive, and robust resource for fish genomics.
The teleost duplication was followed by delayed rediploidization
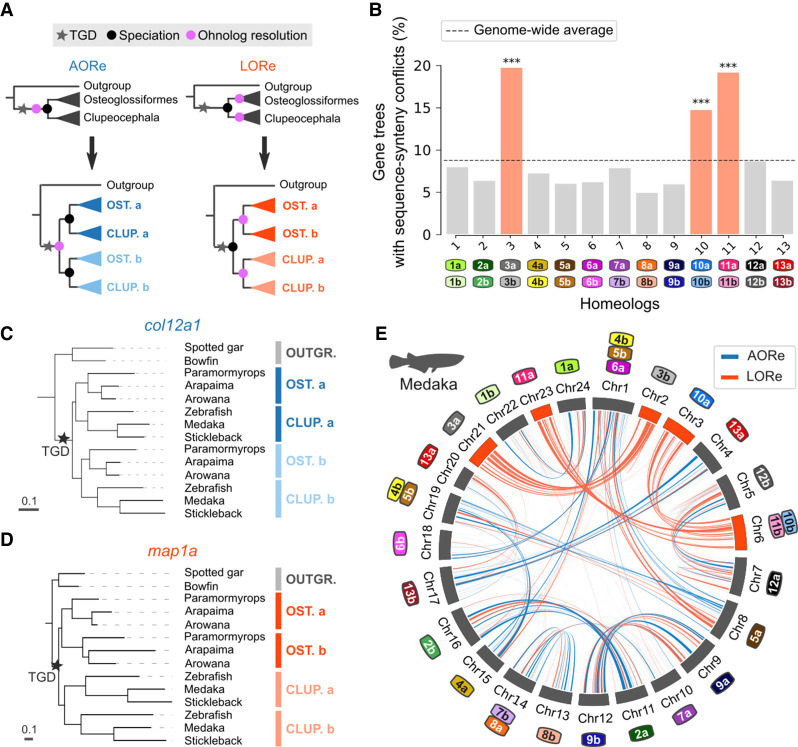
The comparative genomic atlas is the first resource that allows an in-depth, genome-wide analysis of the TGD, and in particular of the early genome evolution processes that followed after the TGD. Previous work has revealed that auto- and allotetraploids significantly differ in their early evolution (Stebbins 1947; Garsmeur et al. 2014; Mason and Wendel 2020): specifically, autotetraploids initially harbor four near-identical chromosome sets, allowing multivalent meiotic recombination and tetrasomic inheritance to occur until preferential bivalent pairings evolve (i.e., cytological rediploidization). Previous studies in salmonids and Acipenseriformes have revealed that this process is highly dynamic, occurring in distinct temporal waves for different genomic regions, separated by as much as tens of millions of years (Robertson et al. 2017; Gundappa et al. 2022; Redmond et al. 2022). Sequence exchanges caused by prolonged multivalent recombination are detectable because they delay the divergence of duplicated regions until after cytological rediploidization occurs, resulting in different phylogenetic expectations regarding ohnolog sequence evolution. Depending on the respective timings of rediploidization and speciation, ohnologs can either follow the ancestral ohnolog resolution (AORe) or lineage-specific ohnolog resolution (LORe) models (Fig. 2A), introduced in Robertson et al. (2017). In the AORe model, cytological rediploidization occurs before lineages split, thus initiating ohnologous sequence divergence before speciation. In the LORe model, because cytological rediploidization has not been resolved before speciation occurs, ohnologs share more sequence similarity within clades than across clades, and can therefore be misidentified as clade-specific duplications.
Figure 2.
Delayed rediploidization following the TGD. (A) Gene tree topologies expected under the AORe and LORe models. The AORe tree topology assumes that rediploidization was complete before the divergence of Osteoglossiformes and Clupeocephala, initiating “a” and “b” gene sequence divergence before speciation. The LORe tree topology assumes that rediploidization was completed only after the divergence of Osteoglossiformes and Clupeocephala, delaying “a” and “b” duplicated sequence divergence to after speciation. (B) Ancestral Chromosomes 3, 10, and 11 are enriched in sequence-synteny conflicts (Methods, [***] P < 0.001, hypergeometric tests with Benjamini–Hochberg correction for multiple testing). Color labels identify ancestrally duplicated chromosomes as in Figure 1B. (C) Examples of an AORe gene tree. For the col12a1a - col12a1b family, the LORe topology is inconsistent with gene sequence evolution (P = 4 × 10−9, AU-test). (D) Example of a LORe gene tree. For the map1aa - map1ab family, AORe was rejected (P-value = 0.001, AU-test). (E) AORe and LORe gene families visualized on the medaka karyotype. Medaka chromosomes are annotated as numbers, whereas color labels represent ancestral chromosomes (Methods), as in (B). Homeologs 3, 10, and 11 almost entirely rediploidized later than the Osteoglossiformes/Clupeocephala divergence.
To investigate the polyploidization mode and rediploidization processes of teleost fishes, we established whether meiosis was fully resolved with bivalent chromosomal pairing in the teleost ancestor before extant teleost lineages diverged, or whether some duplicated genomic regions still recombined during meiosis at that time. We focus our analysis at the Osteoglossiformes/Clupeocephala lineage split, which is the earliest possible speciation point in the teleost phylogeny (Parey et al. 2022), dated approximately at 267 Mya (Kumar et al. 2017). We selected a subset of six teleost genomes (Paramormyrops, arapaima, Asian arowana, zebrafish, medaka, and stickleback) to ensure an equal representation of Osteoglossiformes and Clupeocephala genomes in the data set, along with outgroup genomes (Methods; Supplemental Fig. S5). We developed an extension to SCORPiOs named LORelEi for “Lineage-specific Ohnolog Resolution Extension”. SCORPiOs LORelEi is built around two modules (“diagnostic” and “likelihood tests”) to identify delayed rediploidization in gene trees. The LORelEi diagnostic module leverages the gene tree correction applied by SCORPiOs to identify sequence-synteny conflicts. Sequence-synteny conflicts arise when the orthology relationships of a gene family are inconsistent with conserved synteny information, but correcting the WGD duplication node to rectify this inconsistency induces a significant drop in likelihood estimated from sequence divergence. We grouped gene trees with sequence-synteny conflicts according to their ancestral chromosome of origin in the comparative atlas. Three anciently duplicated chromosome pairs (3, 10, and 11) are significantly enriched in sequence-synteny conflicts, thus hinting toward prolonged recombination between these homeologs after the TGD (Fig. 2B; P < 0.05, hypergeometric tests corrected for multiple testing).
We next sought to confirm that the identified sequence-synteny conflicts were consistent with the phylogenetic expectations of delayed rediploidization. We used the LORelEi likelihood test module to explicitly compare the likelihoods for the tree topologies expected under the AORe and LORe rediploidization models, in which ohnolog resolution occurs before or after the Osteoglossiformes/Clupeocephala lineage split, respectively (Fig. 2A; Methods). We performed these likelihood tests on 5557 gene trees, which retain both ohnologs in at least one of the descending lineages. For 638 gene trees, the early resolution topology (AORe) was significantly more likely, whereas the late resolution topology (LORe) was favored for 1361 trees (likelihood AU-tests, alternative topology rejected at α = 0.05; no significant differences for the remaining 3558 trees). For example, the col12a1a - col12a1b ohnologs had stopped recombining and accumulated independent substitutions by the time Osteoglossiformes and Clupeocephala diverged (Fig. 2C). On the other hand, the map1aa - map1ab TGD ohnologs presumably still recombined when the taxa diverged, and both ohnologs diverged independently later on in each lineage (Fig. 2D). We mapped the location of these genes on the medaka karyotype, which remains close to the ancestral teleost karyotype, to identify chromosomal regions of ancestral and lineage-specific rediploidization (Fig. 2E; Methods). This visualization revealed that ancestral chromosome pairs 3, 10, and 11 had not yet rediploidized when teleosts started diversifying, with large runs of LORe ohnologous families spanning the entire length of their descendant chromosomes in medaka (Chromosomes 2, 3, 6, 21, and 23). The other homeologs appear as a mix of localized AORe and LORe regions, suggesting that rediploidization was ongoing.
This snapshot of the rediploidization status at the time of the Osteoglossiformes/Clupeocephala lineage split shows that rediploidization was not uniform across the ancestral teleost genome, consistent with results in salmonids (Robertson et al. 2017; Gundappa et al. 2022). In salmonids, different temporal waves of rediploidization have been linked with variations in gene functions (Gundappa et al. 2022). In teleosts, we found no evidence of functional enrichments in either the AORe genes, which rediploidized early, or the LORe genes carried by late-rediploidized ancestral Chromosomes 3, 10, and 11. Interspersed LORe genes on other homeologs were functionally enriched for different pathways, including “TGF-beta signaling pathway,” “regulation of transferase activity,” and “regulation of intracellular signal transduction” (corrected P-values < 0.05; Supplemental Tables S3, S4; Methods). One possible explanation to the functional differences between the two sets of LORe genes might be that homeologous Chromosomes 3, 10, and 11 remained tetraploid owing to specificities in their nuclear organization and topological features, which prevented rediploidization, whereas sequence exchanges at LORe loci on other homeologs may have been locally maintained as a result of selection, for example by removing deleterious mutations or transferring favorable alleles at all gene loci. In this speculative model, meiosis resolution would largely occur in waves because some chromosomes or chromosomal regions are mechanistically “easier” to rediploidize than others.
Altogether, we provide the first evidence that entire chromosomes experienced delayed rediploidization in teleosts and continued to exchange genetic material between homeologs for at least 60 million years after the teleost whole genome duplication. This prolonged exchange of genetic material between duplicated chromosomes after the TGD strongly suggests that the teleost ancestor was an autotetraploid.
The teleost genome experienced biased gene retention after duplication
Previous observations on a limited subset of genes have suggested that the ancestral teleost genome has experienced biased gene retention after the TGD (Conant 2020). Biased gene retention is generally considered a hallmark of allopolyploidization (Garsmeur et al. 2014), but has mostly been observed in plants, and the mechanisms of gene retention in polyploid vertebrate genomes may differ. We tested whether the ancestral tetraploid teleost underwent biased gene retention, with one duplicated chromosome copy systematically retaining more genes than the other. We computed gene retention in nonoverlapping 10-gene windows along duplicated chromosome copies in several teleost genomes, using an outgroup fish as proxy for the ancestral gene order (Fig. 3A,B; Supplemental Figs. S6, S7; Methods) and total gene retention on homeologs in the ancestor (Osteoglossocephalai) of all 74 teleosts (Supplemental Table S5). We uncover a consistent, significant bias in gene retention on ancestral chromosome pairs 3, 4, 7, 11, and 13, in which genes were preferentially retained on one homeolog over the other, regardless of study species. We find additional but weaker evidence for biased gene retention on ancestral chromosome pairs 2, 5, 6, 8, and 9 in some combinations of genome comparisons. We however do not find evidence of retention bias between ancestrally duplicated copies for chromosome pairs 1, 10, and 12. This preferential gene retention is not an artifact of the atlas construction because of genes unassigned to either duplicated chromosome: indeed, conservatively assigning such genes to the homeolog with the lowest retention did not offset the retention imbalance (Supplemental Table S6). In summary, we find that genes were preferentially retained on one duplicated chromosome in a subset of ancestral chromosomes, a character more frequently observed in allopolyploids (Garsmeur et al. 2014; Cheng et al. 2018). Of note, the chromosome copy with highest gene retention is typically annotated as “a” in the atlas as a consequence of the construction process. As the TGD is likely an autopolyploidization event, “a” and “b” correspond to different chromosome copies and are interchangeable for each chromosome pair—“a” and “b” chromosomes should not be interpreted as distinct parental subgenomes of allopolyploidization.
Figure 3.
Differences in gene retention, selective pressure, and gene expression between duplicated chromosomes. (A) Schematic example of gene retention calculation. Using an outgroup genome as an approximation of the ancestral gene order, we assess gene retention on each duplicated chromosome in teleost genomes, by 10-gene bins, regardless of their genomic location (Methods). (B) Gene retention on anciently duplicated chromosome copies in medaka, using the spotted gar genome as a proxy for ancestral gene order. Ancestral chromosomes with a significant bias in gene retention on one of the two copies are highlighted ([***] P < 0.001, [**] P < 0.01, [*] P < 0.05, Wilcoxon paired tests with Benjamini–Hochberg correction for multiple testing). (C) Number of genes experiencing relaxed selection compared with their ohnolog across homeologs (Methods; Fisher's exact tests with Benjamini–Hochberg correction for multiple testing, P-values as in B). (D) Average expression across tissues in medaka. No significant differences in expression were detected between genes of duplicated chromosome copies (Wilcoxon paired tests with Benjamini–Hochberg correction for multiple testing, at α = 0.05).
Differences in selective pressures and gene expression do not explain the observed bias in gene retention
Previous observations have suggested that biased gene retention in allopolyploid genomes reflects partial epigenetic silencing of one parental subgenome (Freeling et al. 2012; Bird et al. 2021). Genes on silenced chromosome copies are expressed at lower levels, and therefore subjected to lower selective pressures and ultimately to pseudogenization. We therefore investigated whether anciently duplicated chromosome copies that retained fewer genes have decayed because of unequal selective pressure. We estimated dN/dS ratios for 1263 pairs of TGD-derived paralogs conserved in at least 40 species, and tested whether ohnologs on one of the two anciently duplicated chromosomes systematically underwent more relaxed sequence evolution (Methods). On ancestral chromosome pairs 8 and 13, genes of one chromosome copy experienced a significant relaxation of selection compared with their ohnologous counterparts (Fig. 3C). Genes under relaxed selection were located on the homeolog with lower retention, as predicted if low-selective pressure promotes gene loss. However, homeologs with significant differences in selective pressure correspond only to two out of six chromosomes showing strongly biased gene retention (n = 13; P = 0.1923; Fisher's exact test), with nonsignificant differences in selective pressure in the same direction as the gene retention bias for two chromosome pairs (4 and 11) and in the opposite direction for the two others (3 and 7). Thus, differences in selective pressures do not explain the observed bias in gene retention.
In addition, we investigated whether ancestral chromosome copies show differences in gene expression, which would have been established after the TGD and have been maintained since. We find no bias in ohnolog gene expression between ancestral chromosomes from each pair. This result is consistent whether looking at average gene expression across 11 tissues in medaka (Fig. 3D), zebrafish (Supplemental Fig. S8), or tissue by tissue in either medaka or zebrafish (Supplemental Figs. S9, S10).
In conclusion, our findings are consistent with previous reports that gene retention on ancestral chromosomes was biased following the teleost duplication (Conant 2020). However, we find that biased gene retention in teleosts is not correlated with other features classically observed in allopolyploids, and our results suggest that biased gene retention can occur following autopolyploidization, possibly driven by distinct and underappreciated factors.
The comparative atlas enhances teleost gene and genome annotations
Finally, we investigated how the comparative atlas may improve crucial resources for fish evolutionary, ecological, and functional genomics. The Zebrafish Information Network (ZFIN) provides manually curated, high-quality reference annotations for zebrafish genes and implements rigorous conventions for gene naming (Ruzicka et al. 2019). These gene names are then propagated to orthologous genes in other teleost genomes, providing the basis of the entire teleost gene nomenclature and functional annotation transfers. In an effort to convey evolutionary meaning within the gene names, zebrafish paralogs descended from the TGD are identified with an “a” or “b” suffix. ZFIN guidelines recommend that adjacent genes should carry the same suffix when they belong to a continuous syntenic block inherited since the TGD, sometimes called syntelogs (Zhao et al. 2017). This aspiration has however been difficult to implement in the absence of a high-resolution map of zebrafish duplicated regions and their ancestral chromosomes of origin. To assess the consistency in consecutive zebrafish gene names, we extracted and compared zebrafish “a” and “b” gene suffixes along duplicated regions from the comparative atlas (Fig. 4). We find that despite previously mentioned efforts, zebrafish “a” and “b” gene suffixes are not consistent with the polyploid history of the zebrafish genome (Fig. 4A): 43% of gene suffixes would have to be reassigned in order to reflect the shared history of syntenic genes, which would be impractical to implement (Methods). Gene annotations are therefore unhelpful to study large-scale processes such as chromosome evolution or genome-wide rediploidization. Additionally, the ZFIN nomenclature does not impart a suffix to singleton genes, which correspond to TGD-duplicated genes in which one of the copies was eventually lost. As a result, only 26% of zebrafish genes are annotated with an “a” or “b” suffix in ZFIN.
Figure 4.
Zebrafish gene names are not evolutionarily consistent. (A) Karyotypic localization of zebrafish “a” and “b” TGD ohnologs, according to the ZFIN annotation. ZFIN does not annotate genes as either “a” or “b” when one of the TGD paralogs has been lost, and these genes are not represented here. (B) Complementary annotation of zebrafish “a” and “b” gene copies using the comparative atlas (84% of zebrafish genes annotated, including genes without a TGD ohnolog).
Using the comparative atlas, we annotated which paralogs were retained for 84% of all zebrafish genes, providing a complementary resource that significantly extends the evolutionary annotation of gene histories in this species (Fig. 4B). The comparative atlas also includes similar annotations for all 74 studied teleosts, as shown for medaka, stickleback, and tetraodon (Supplemental Fig. S11). How ancient tetraploids return to a mostly diploid state is an active area of research, where distinguishing which paralog gene has been retained can be essential (Inoue et al. 2015; Robertson et al. 2017; Conant 2020; Simakov et al. 2020; Gundappa et al. 2022). The comparative atlas opens the possibility to formally identify and investigate which ancestral copies have been retained and lost during teleost diversification, and transfer functional gene annotations between model and nonmodel fish genomes. As such, the comparative atlas constitutes a biologically meaningful, historically accurate insight into reference gene annotations to support further investigations of teleost genome evolution.
Discussion
Teleost fishes have a long-standing history as tractable model species for vertebrate development and human disease (Ohno et al. 1968; Streisinger et al. 1981; Haffter et al. 1996; Lieschke and Currie 2007; Schartl 2014), and have contributed major breakthroughs in ecological, evolutionary, and functional genomics over the years (Braasch et al. 2016; Capel 2017; Xie et al. 2019). Teleosts have experienced several whole-genome duplications during their diversification, a process which has been essential to early vertebrate evolution. Vertebrates underwent two foundational WGDs more than 450 million years ago (Dehal and Boore 2005), and these events have contributed to the establishment of the vertebrate karyotype as we know it (Sacerdot et al. 2018; Simakov et al. 2020), as well as to the expansion of major gene families such as the Hox clusters and MHC genes (Castro et al. 2004; Dehal and Boore 2005). As WGDs are rarer in vertebrates than they are in plants, our knowledge regarding post-WGD evolution is anchored on processes observed in plants, in which these events are frequent and mechanistically diverse. However, functional constraints on genome and organismal evolution are significantly different between plants and vertebrates. How principles of plant polyploid evolution extend to vertebrates is not well understood, and characterizing ancient WGD events such as the teleost whole-genome duplication is essential to illuminate and interpret the early genetic mechanisms at the origin of vertebrates.
One long-standing question with respect to the teleost duplication is whether the tetraploid teleost ancestor arose via allopolyploidization or autopolyploidization (Martin and Holland 2014; Conant 2020). Previous studies have highlighted the contrasting genomic implications of allopolyploidization and autopolyploidization, in which the two subgenomes of allopolyploids are often—although not systematically—subjected to asymmetrical evolution, whereas in autopolyploids all chromosome copies are highly similar and equally affected by the rediploidization process (Garsmeur et al. 2014; Cheng et al. 2018). We leveraged the comparative atlas to investigate how the postduplication rediploidization process has shaped extant teleost genomes and reveals the early history of tetraploid fishes. We find evidence for prolonged recombination between entire duplicated chromosomes after the TGD, thus strongly suggesting that the ancestor of teleosts was an autopolyploid. Our results provide the first genome-wide support for delayed rediploidization in teleosts following the TGD, previously suggested as one potential explanation for the enigmatic evolutionary history of the Hox and rhodopsin genes in teleosts, and of several gene families in eels (Martin and Holland 2014; Nakamura et al. 2017; Rozenfeld et al. 2019). We show that delayed rediploidization after the Osteoglossiformes/Clupeocephala lineage split did not only affect these specific gene segments but do extend to entire ancestral chromosomes.
In addition, we find a significant bias in gene retention for a subset of duplicated chromosome pairs, as also previously observed at a more local scale (Conant 2020). We lack an explanatory mechanism for these differences, which have been classically linked to allopolyploid genome rediploidization, in which one parental subgenome is more expressed, under stronger selective pressure, and retains more genes. Here, we find no clear correlation linking together differences in gene retention, expression level, and selective pressure. Although the well-studied examples of salmonid and carp genome duplications recapitulate classical models of autopolyploid and allopolyploid rediploidization, respectively (Robertson et al. 2017; Xu et al. 2019; Gundappa et al. 2022; Li et al. 2021), polyploid karyotype evolution in vertebrates can also clearly be more complex and involve additional, less appreciated factors that remain to be investigated. We highlight that biased gene retention cannot be considered as reliable evidence in favor of allopolyploidization in vertebrates, as we provide formal evidence that the same chromosomes can initially recombine and then experience biased gene retention, suggesting that this bias is unrelated to initial sequence divergence in teleosts.
These novel insights into the rediploidization processes of teleost genomes have overarching implications for fish evolutionary genomics. First, as the ancestral teleost was a probable autotetraploid, the formal distinction between ancestral subgenomes is irrelevant, and the annotations of chromosomal copies in the comparative atlas should not be misinterpreted as two distinct parental subsets, in which all “a” chromosomes (or “b”) descend from a single parental genome. Second, delayed rediploidization has profound consequences on the dynamics of gene evolution. After whole-genome duplication, duplicated gene copies can diverge and undergo specialized evolution by partitioning ancestral functions (subfunctionalization) or acquiring new expression patterns (neofunctionalization) (Ohno et al. 1968; Force et al. 1999; Lynch and Conery 2000). These processes are thought to contribute to diversification and the acquisition of lineage-specific traits: therefore, resolving gene orthology relationships between species is critical to investigate the dynamics of ohnolog gene retention, divergence and loss and their involvement in phenotypic novelty. However, in this evolutionary scenario, ohnolog sequences only start diverging once recombination is suppressed, which implies that there are no strict 1:1 orthology relationships between duplicated genes across species that separated before meiosis resolution. As such, “a” and “b” gene assignments are not informative of the underlying sequence information contained in genomic regions that showed delayed rediploidization, and these genes should be considered as tetralogs rather than paralogs and orthologs (Martin and Holland 2014; Robertson et al. 2017). The genetic and functional divergence of these genes has occurred independently in each subsequent lineage, and further inquiry will reveal whether these specific evolutionary dynamics have contributed to lineage-specific evolution in the major teleost clades, as described in salmonids (Robertson et al. 2017; Gundappa et al. 2022). In particular, further work across teleosts and other (paleo)polyploid clades are required to untangle the contributions of mechanistic constraints to cytological rediploidization dynamics, and the evolutionary drivers delaying or promoting the genetic divergence of ohnologs.
It should be noted that the teleost comparative atlas comes with a number of limitations that directly stem from the way it was built. First, the assignations to ancestral chromosomes before the TGD are only as good as the state-of-the-art ancestral genome reconstruction. There is a general consensus that the ancestral teleost genome contained 13 chromosomes (Kasahara et al. 2007; Nakatani and McLysaght 2017)—however, the precise delineation of genes descending from each of those 13 groups may be subject to modifications as the field evolves, which may lead to updates in the ancestral assignations of the regions in the comparative atlas. Second, although we show that the comparative atlas is resilient to species-specific errors in ancestral chromosome or gene orthology group assignations owing to the redundant information provided by multiple species, the atlas is ultimately based on gene family tree models, which are sometimes inaccurate or incomplete (Hahn 2007; Som 2015). Inaccurate trees may result in local misspecifications of gene paralogs or ancestral assignations in the comparative atlas. We have previously shown that SCORPiOs significantly improves gene tree accuracy after a WGD event (Parey et al. 2020), and we found only few discrepancies between megabase-scale regions predicted to descend from the same ancestral chromosome from Nakatani and McLysaght (2017) and paralogy relationships predicted at gene-to-gene resolution by our gene trees. This suggests that the atlas is generally accurate. However, we note that SCORPiOs flagged 2832 gene trees in which sequence identity relationships are inconsistent with their local syntenic context, which may represent areas in which the comparative atlas is either less reliable or biologically less informative. To conclude, as teleost fishes have become a high priority taxon for several large-scale projects aiming to extend phylogenetic coverage of vertebrate genome resources (Fan et al. 2020; Rhie et al. 2021), we expect that the genome-scale, clade-wide paralogy and orthology resources we provide here will propel the functional and evolutionary characterization of this major clade encompassing more than half of all vertebrate species.
Methods
Libraries and packages
Scripts to build the comparative atlas were written in Python 3.6.8 and assembled together in a pipeline with Snakemake version 5.13.0 (Köster and Rahmann 2012). The ete3 package (version 3.1.1) was used for phylogenetic gene tree manipulation and drawing. Other Python package dependencies used for plots and analyses include Matplotlib (version 3.1.1), seaborn (version 0.9.0), numpy (version 1.18.4), pandas (version 0.24.2), pingouin (version 0.3.4) for Wilcoxon paired tests and multiple testing correction, and SciPy (version 1.4.1) for Fisher's exact tests. Chromosome painting and synteny comparisons were drawn with the RIdeogram R package (Hao et al. 2020).
Genomic resources
Genome assemblies and annotations for the 101 vertebrate genomes were downloaded from various sources, including the NCBI, Ensembl version 95 and GigaDB. The source and assembly version used for each genome are listed in Supplemental Table S1. The gene coordinates files for the 74 teleost genomes in the comparative atlas have been deposited to Zenodo (https://doi.org/10.5281/zenodo.5776772).
Phylogenetic gene trees
Initial gene trees were built using the Ensembl Compara pipeline (Vilella et al. 2009). Briefly, starting from the sets of proteins derived from the longest transcripts in each genome, we performed an all-against-all BLASTP+ (Altschul et al. 1990), with the following parameters “-seg no -max_hsps 1 -use_sw_tback -evalue 1e-5”. We then performed clustering with hcluster_sg to define gene families, using parameters “-m 750 -w 0 -s 0.34 –O”. We built multiple alignments using M-Coffee (Wallace et al. 2006), with the command “t-coffee -type = PROTEIN -method mafftgins_msa, muscle_msa,kalign_msa,t_coffee_msa -mode = mcoffee”. Next, we conducted phylogenetic tree construction and reconciliation with TreeBeST, using default parameters (Vilella et al. 2009). Although TreeBeST remains the most efficient method to build gene trees for large data sets (Noutahi et al. 2016), it systematically infers a number of gene duplication nodes that are overly old and poorly supported (Hahn 2007). We therefore edited the TreeBeST gene trees: we used ProfileNJ (Noutahi et al. 2016) to correct nodes with a very low duplication confidence score (duplication score < 0.1, computed as the fraction of species that retained the genes in two copies after the duplication). Specifically, ProfileNJ rearranges subtrees below these poorly supported nodes to make them more parsimonious in terms of inferred duplications and losses. Finally, we ran SCORPiOs (version v1.1.0) to account for several whole-genome duplication events in the species phylogeny and correct gene trees accordingly: the teleost duplication (TGD), using bowfin and gar as outgroups. SCORPiOs reached convergence after five iterative rounds of correction. We identified 8144 teleost gene subtrees out of 17,493 (47%) that were inconsistent with synteny information, of which 5611 could be corrected (32% of all subtrees). We note that the corrected-to-inconsistent tree ratio (69%) is comparable to our previous application of SCORPiOs to fish data. Similarly, the proportion of errors in initial sequence-based gene trees is in line with our previous application of SCORPiOs to a data set of 47 teleost species (Parey et al. 2020). We also applied SCORPiOs to correct the nodes corresponding to the carp 4R WGD, using zebrafish as outgroup; and the salmonid 4R WGD, using Northern pike as outgroup. In the presence of LORe in salmonids, as is the case for teleosts, SCORPiOs will attempt to correct gene trees on the basis of synteny information but the correction will be rejected for being inconsistent with sequence evolution. As a result, the salmonid 4R WGD will be placed as lineage-specific duplications in LORe trees, consistent with sequence evolution.
Ancestral gene statistics
We calculated the predicted number of genes in the postduplication ancestral teleost genome using our set of 26,692 gene trees, and compared this to 60,447 state-of-the art gene trees stored in Ensembl Compara v100. Specifically, to calculate the number of genes inferred in the teleost ancestor (Osteoglossocephalai), we counted ancestral gene copies assigned to Osteoglossocephalai in the 26,692 and 60,447 TreeBeST phylogeny-reconciled gene trees. This ancestral gene number is an indirect but accurate approximation of the quality of inferred gene trees, because the major challenge is to accurately position duplications at this ancestral node.
Comparative genomic atlas
The FishComparativeAtlas pipeline annotates teleost genes to post-TGD duplicated chromosomes. It takes as input genomic regions annotated to preduplication chromosomes for four reference species (zebrafish, medaka, stickleback, and tetraodon), and the set of gene trees described above. Segmentation of the four teleost species with respect to the 13 ancestral chromosomes were extracted from Nakatani and McLysaght (2017) and genomic interval coordinates converted to lists of genes. All genomes were then reduced to ordered lists of genes. We first converted input genomic intervals from zebrafish genome assembly Zv9 to GRCz11 and from medaka genome assembly MEDAKA1 to ASM223467v1, by transferring boundary genes between assemblies using Ensembl gene ID histories. Next, we identified putative TGD sister regions within each of the four reference species, as regions sharing a high fraction of TGD-descended paralogs (Supplemental Fig. S2B). We grouped regions descended from the same preduplication ancestral chromosome, and iteratively annotated pairs of regions into internally consistent sets of “a” and “b” postduplication sister regions as follows: for each ancestral chromosome, we started from the largest descendant region and arbitrarily defined it as “a”. All regions sharing 50% ohnologs or more with this “a” region are identified as the “b” paralog region(s). Additional search rounds were then conducted to extend the “a” and “b” annotations in a stepwise manner to all regions descended from this ancestral chromosome. The required ohnolog fraction was decreased at each round, and the search was stopped when remaining blocks shared <5% ohnologs with previously annotated regions. Because this identification of duplicated regions was performed independently in each of the four species, “a” and “b” region annotations were homogenized to ensure consistency across species (Supplemental Fig. S2C). The homogenization step uses orthology relationships from the gene trees and stickleback as an arbitrary guide species: annotations were switched in other species when “a” segments shared more orthologous genes with the “b” region of stickleback. Where conflicts remained for individual gene annotations, we used a majority vote procedure across all four species to define the postduplication chromosome. Finally, annotations were propagated to all teleost genomes in the gene trees using orthologies.
Simulation of ancestral chromosome boundary shifts
To simulate uncertainty in interval boundaries in the original ancestral genome reconstruction from Nakatani and McLysaght (2017), we randomly drew new boundaries in the vicinity of their original location according to a Gaussian distribution with standard deviation σ varying in [5, 10, 25, 50, 75, 100] genes. These boundary shifts were independently generated for each of the four reference species, with n = 100 random noise simulations for each σ value and each reference species. In total, simulations generated 600 noisy input data sets that were processed with the FishComparativeAtlas pipeline to assess its robustness to noise. Each of the 600 produced outputs were then compared to the comparative atlas, by counting the proportion of gene families with a reassigned ancestral chromosome (Supplemental Fig. S3).
Early and late rediploidization gene tree topologies
Phylogenetic gene trees were built for the reduced set of 33 genomes as follows. We first filtered the CDS multiple alignments previously computed for the 74 teleosts and 27 nonteleost outgroup genomes set as described above, to retain only genes of all 27 outgroups and 6 teleost genomes, including three Osteoglossiformes: paramormyrops (Paramormyrops kingsleyae), arapaima (Arapaima gigas), and arowana (Scleropages formosus) and three Clupeocephala: zebrafish (Danio rerio), medaka (Oryzias latipes), and stickleback (Gasterosteus aculeatus) (Supplemental Fig. S5). We used these reduced multiple alignments to build phylogenetic gene trees with TreeBeST (Vilella et al. 2009), using default parameters and the option “-X 10”. This resulted in a set of 14,391 gene trees containing genes of the 33 retained genomes. We then ran SCORPiOs to correct trees for the teleost-specific duplication using the gar and bowfin genomes as outgroups.
To investigate the occurrence of delayed rediploidization, we implemented an extension to the SCORPiOs pipeline (SCORPiOs “LORelEi” for “Lineage-specific Ohnolog Resolution Extension”) to (1) identify gene trees characterized by sequence-synteny prediction conflicts and (2) perform likelihood AU-tests (Shimodaira and Hasegawa 2001) to evaluate how AORe and LORe rediploidization tree topologies are supported by gene sequence evolution. For (1), we identify gene trees that SCORPiOs attempts to correct based on syntenic information, but whose correction is rejected because of low sequence-based likelihood. For (2), we selected the 5557 gene families containing a gene in at least one reference outgroup (bowfin or gar) and resulting in distinct tree topologies under AORe and LORe. In practice, these topologies can be distinguished when at least one teleost group (Osteoglossiformes or Clupeocephala) retained both ohnologs, although not necessarily in the same species. For each of these 5557 families, we built three gene trees using RAxML 8.2.12 (Stamatakis 2014), with the GTRGAMMA model: the unconstrained maximum likelihood (ML) tree, the constrained AORe topology, and the constrained LORe topology (Fig. 2B). We then used CONSEL (Shimodaira and Hasegawa 2001) to test for significant differences in log-likelihood reported by RAxML (Stamatakis 2014). A tree topology was rejected when significantly less likely than the ML tree at α = 0.05.
We used Circos version 0.69–9 (Krzywinski et al. 2009) to visualize AORe and LORe ohnologs on the medaka genome. Before the Circos construction, we used the “bundlelinks” tool available in the circos-tools suite version 0.23 to bundle together consecutive genes with the same rediploidization mode, using 50 genes as the maximum distance parameter (-max_gap 50). Using this setting, isolated AORe and LORe are less visible (i.e., have thinner links) than high-confidence regions of consecutive genes displaying the same rediploidization mode. On the Circos, we annotate ancestral chromosomes corresponding to each medaka chromosome with color labels. For clarity purposes, we only add a label if more than 17.5% of genes of a given medaka chromosome are annotated to the ancestral chromosome.
Functional enrichment tests
For each of the AORe, LORe whole-chromosomes, and LORe interspersed sets, we extracted high-confidence ohnologs list in medaka. Specifically, we retained only ohnologs falling in high-confidence AORe and LORe regions defined by “bundled” gene families in the Circos representation (i.e., regions formed from neighboring genes with the same rediploidization mode, using 50-gene sized windows). Finally, we used the zebrafish orthologs of these genes (n = 248 zebrafish genes for AORe, n = 193 for LORe whole-chromosomes and n = 215 for LORe interspersed) to perform Gene Ontology and KEGG pathway enrichment analyses through the WebGestalt web server (Liao et al. 2019).
Gene retention on homeologous chromosomes
Because ancestral gene order is particularly difficult to reconstruct in teleosts because of an elevated rate of microsyntenic rearrangements and gene copy losses (Inoue et al. 2015; Nakatani and McLysaght 2017), we take advantage of a nonduplicated outgroup fish genome as a proxy for the ancestral gene order. Here, we make the assumption that consecutive genes on the outgroup genome, all assigned to the same preduplication chromosome, represent the ancestral gene order. Using the gene trees, we identify 10,629 1-to-1 orthologies between spotted gar genes and teleost preduplication gene families, and 11,599 with bowfin genes. We then used the gene order in the outgroup genomes as an approximation of the ancestral teleost gene order. We reduced outgroup genomes to these genes, and extracted all blocks of consecutive genes annotated to the same preduplication chromosome. We computed the percentage of gene copies retained on “a” and “b” homeologs in each extant duplicated genome, using nonoverlapping windows of 10 genes along these blocks.
Tests for relaxed selective pressures
We considered 1263 teleost gene families annotated in the comparative atlas for the dN/dS analysis, selecting all families that contained exactly two ohnologous copies in at least 40 teleost genomes (excluding salmonids and carps, which underwent additional WGDs), and exactly one orthologous copy in the bowfin and gar outgroups. We pruned the trees from any species with only one ohnolog copy or where additional gene duplications were present, to obtain “a” and “b” clades with the exact same species, for informative dN/dS comparisons. For each gene family, we used translatorX vLocal (Abascal et al. 2010) to (1) translate coding sequences, (2) align resulting amino acid sequences with MAFFT v7.310 (Katoh and Standley 2013) using option “‐‐auto”, (3) trim poorly aligned regions with Gblocks version 0.91b (Castresana 2000) using parameters “-b4 = 2 -b5 = h”, and (4) back-translate the sequences into codon alignments. We used the RELAX model in HyPhy (Wertheim et al. 2015) to estimate dN/dS ratios and test for significant relaxation or intensification of selection on branches of the “a” subtree compared with branches of the “b” subtree. Briefly, RELAX estimates dN/dS distributions across sites on the sets of “a” and “b” branches and fits a selection intensity parameter “k”, which captures the extent of selection intensification (k > 1) or relaxation (k < 1). Likelihood ratio tests are conducted to compare the alternative model with the k parameter to a null model without. We identified relaxation or intensification of selection on “a” versus “b” branches when the null model was rejected (P-values < 0.05, corrected for multiple testing using the Benjamini–Hochberg procedure). Finally, we performed Fisher's exact tests corrected for multiple testing using the Benjamini–Hochberg procedure, to identify chromosome pairs with significantly higher numbers of genes under relaxed evolution on one chromosomal copy.
Expression level of ohnologous genes
We used RNA-seq data sets across 11 tissues (bones, brain, embryo, gills, heart, intestine, kidney, liver, muscle, ovary, and testis) in zebrafish and medaka from the PhyloFish database (Pasquier et al. 2016), normalized into TPM (transcripts per million transcripts) and quantile normalized across tissues as previously described (Parey et al. 2020). Gene IDs were then converted from Ensembl version 89 to version 95 using conversion tables downloaded from BioMart (Smedley et al. 2009). Average expression across tissue (Fig. 3C; Supplemental Fig. S8) and by-tissue expression (Supplemental Figs. S9, S10) were calculated for ohnologs grouped by their ancestral chromosome of origin.
ZFIN gene names
Zebrafish ZFIN gene names were extracted using BioMart (Smedley et al. 2009) from the Ensembl database (version 95). We extracted the last letter of gene names, which represents “a” and “b” ohnology annotations in ZFIN. We then computed the minimal number of “a” and “b” ZFIN gene name reassignments that would be necessary to be consistent with the comparative atlas. In the comparative atlas, “a” and “b” labels are arbitrarily assigned to duplicated chromosomes; that is, genes descended from Chromosomes 1a and 1b could be swapped to 1b and 1a. To not artificially overestimate discordances, we first swapped such arbitrary “a” and “b” annotations to minimize differences with ZFIN. Finally, we counted the remaining number of “a” and “b” disagreements for zebrafish genes in the comparative atlas that were also annotated in ZFIN.
Software availability
All code for the FishComparativeAtlas pipeline is publicly available at GitHub (https://github.com/DyogenIBENS/FishComparativeAtlas). An archive containing a stable version of the code along with all input data (including gene trees) and the final atlas has been deposited in Zenodo (https://doi.org/10.5281/zenodo.5776772), to reproduce the generation of the comparative atlas or directly inspect it. The SCORPiOs LORelEi extension is available at GitHub (https://github.com/DyogenIBENS/SCORPIOS) and has also been deposited in Zenodo (https://doi.org/10.5281/zenodo.6913688), along with all input data, environments, and outputs. In addition, both the FishComparativeAtlas pipeline and SCORPiOs LORelEi extension are available in the Supplemental Material.
Data access
Gene homology relationships and local synteny conservation between teleosts can be interactively browsed through the Genomicus web server (Nguyen et al. 2022), accessible at https://www.genomicus.bio.ens.psl.eu/genomicus-fish-03.01/cgi-bin/search.pl. The homology relationships from the comparative atlas can be downloaded as flat or HTML files via an ftp server (ftp://ftp.biologie.ens.fr/pub/dyogen/genomicus-fish/03.01/ParalogyMap), along with the gene trees in New Hampshire eXtended (NHX) format (ftp.biologie.ens.fr/pub/dyogen/genomicus-fish/03.01/protein_trees.nhx).
Supplementary Material
Acknowledgments
We thank Pierre Vincens for the coordination of computing resources and all members of the GenoFish consortium for fruitful discussions. We also thank Dan Macqueen and two anonymous reviewers for their critical reading of this manuscript and their helpful suggestions to improve its clarity. This work is funded by Agence Nationale de la Recherche (ANR) GenoFish (grant number ANR-16-CE12-0035-02), and was supported by grants from the French Government and implemented by ANR (ANR–10–LABX– 54 MEMOLIFE and ANR–10–IDEX–0001–02 PSL* Research University). This project received funds from the European Union's Horizon 2020 research and innovation programme under Grant Agreement No. 817923 (AQUA-FAANG).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.276953.122.
Competing interest statement
The authors declare no competing interests.
References
- Abascal F, Zardoya R, Telford MJ. 2010. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res 38: W7–W13. 10.1093/nar/gkq291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215: 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Aparicio S, Chapman J, Stupka E, Putnam N, Chia J-M, Dehal P, Christoffels A, Rash S, Hoon S, Smit A, et al. 2002. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 297: 1301–1310. 10.1126/science.1072104 [DOI] [PubMed] [Google Scholar]
- Bird KA, Niederhuth CE, Ou S, Gehan M, Pires JC, Xiong Z, VanBuren R, Edger PP. 2021. Replaying the evolutionary tape to investigate subgenome dominance in allopolyploid Brassica napus. New Phytol 230: 354–371. 10.1111/nph.17137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomblies K, Jones G, Franklin C, Zickler D, Kleckner N. 2016. The challenge of evolving stable polyploidy: could an increase in “crossover interference distance” play a central role? Chromosoma 125: 287–300. 10.1007/s00412-015-0571-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braasch I, Gehrke AR, Smith JJ, Kawasaki K, Manousaki T, Pasquier J, Amores A, Desvignes T, Batzel P, Catchen J, et al. 2016. The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons. Nat Genet 48: 427–437. 10.1038/ng.3526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capel B. 2017. Vertebrate sex determination: evolutionary plasticity of a fundamental switch. Nat Rev Genet 18: 675–689. 10.1038/nrg.2017.60 [DOI] [PubMed] [Google Scholar]
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17: 540–552. 10.1093/oxfordjournals.molbev.a026334 [DOI] [PubMed] [Google Scholar]
- Castro LFC, Furlong RF, Holland PWH. 2004. An antecedent of the MHC-linked genomic region in amphioxus. Immunogenetics 55: 782–784. 10.1007/s00251-004-0642-9 [DOI] [PubMed] [Google Scholar]
- Cheng F, Wu J, Cai X, Liang J, Freeling M, Wang X. 2018. Gene retention, fractionation and subgenome differences in polyploid plants. Nat Plants 4: 258–268. 10.1038/s41477-018-0136-7 [DOI] [PubMed] [Google Scholar]
- Conant GC. 2020. The lasting after-effects of an ancient polyploidy on the genomes of teleosts. PLoS One 15: e0231356. 10.1371/journal.pone.0231356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehal P, Boore JL. 2005. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol 3: e314. 10.1371/journal.pbio.0030314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar-Camacho D, Carleton KL, Narain DW, Pierotti MER. 2020. Visual pigment evolution in Characiformes: the dynamic interplay of teleost whole-genome duplication, surviving opsins and spectral tuning. Mol Ecol 29: 2234–2253. 10.1111/mec.15474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan G, Song Y, Yang L, Huang X, Zhang S, Zhang M, Yang X, Chang Y, Zhang H, Li Y, et al. 2020. Initial data release and announcement of the 10,000 Fish Genomes Project (Fish10K). GigaScience 9: giaa080. 10.1093/gigascience/giaa080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. 1999. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151: 1531–1545. 10.1093/genetics/151.4.1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M, Woodhouse MR, Subramaniam S, Turco G, Lisch D, Schnable JC. 2012. Fractionation mutagenesis and similar consequences of mechanisms removing dispensable or less-expressed DNA in plants. Curr Opin Plant Biol 15: 131–139. 10.1016/j.pbi.2012.01.015 [DOI] [PubMed] [Google Scholar]
- Garsmeur O, Schnable JC, Almeida A, Jourda C, D'Hont A, Freeling M. 2014. Two evolutionarily distinct classes of paleopolyploidy. Mol Biol Evol 31: 448–454. 10.1093/molbev/mst230 [DOI] [PubMed] [Google Scholar]
- Greenway R, Barts N, Henpita C, Brown AP, Rodríguez LA, Peña CMR, Arndt S, Lau GY, Murphy MP, Wu L, et al. 2020. Convergent evolution of conserved mitochondrial pathways underlies repeated adaptation to extreme environments. Proc Natl Acad Sci 117: 16424–16430. 10.1073/pnas.2004223117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundappa MK, To T-H, Grønvold L, Martin SAM, Lien S, Geist J, Hazlerigg D, Sandve SR, Macqueen DJ. 2022. Genome-wide reconstruction of rediploidization following autopolyploidization across one hundred million years of salmonid evolution. Mol Biol Evol 39: msab310. 10.1093/molbev/msab310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffter P, Granato M, Brand M, Mullins MC, Hammerschmidt M, Kane DA, Odenthal J, Kelsh RN, Furutani-Seiki M, Vogelsang E, et al. 1996. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development 123: 1–36. 10.1242/dev.123.1.1 [DOI] [PubMed] [Google Scholar]
- Hahn MW. 2007. Bias in phylogenetic tree reconciliation methods: implications for vertebrate genome evolution. Genome Biol 8: R141. 10.1186/gb-2007-8-7-r141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Z, Lv D, Ge Y, Shi J, Weijers D, Yu G, Chen J. 2020. RIdeogram: drawing SVG graphics to visualize and map genome-wide data on the idiograms. PeerJ Comput Sci 6: e251. 10.7717/peerj-cs.251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero J, Muffato M, Beal K, Fitzgerald S, Gordon L, Pignatelli M, Vilella AJ, Searle SMJ, Amode R, Brent S, et al. 2016. Ensembl comparative genomics resources. Database (Oxford) 2016: bav096. 10.1093/database/bav096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, Muffato M, Collins JE, Humphray S, McLaren K, Matthews L, et al. 2013. The zebrafish reference genome sequence and its relationship to the human genome. Nature 496: 498–503. 10.1038/nature12111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue J, Sato Y, Sinclair R, Tsukamoto K, Nishida M. 2015. Rapid genome reshaping by multiple-gene loss after whole-genome duplication in teleost fish suggested by mathematical modeling. Proc Natl Acad Sci 112: 14918–14923. 10.1073/pnas.1507669112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillon O, Aury J-M, Brunet F, Petit J-L, Stange-Thomann N, Mauceli E, Bouneau L, Fischer C, Ozouf-Costaz C, Bernot A, et al. 2004. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 431: 946–957. 10.1038/nature03025 [DOI] [PubMed] [Google Scholar]
- Kasahara M, Naruse K, Sasaki S, Nakatani Y, Qu W, Ahsan B, Yamada T, Nagayasu Y, Doi K, Kasai Y, et al. 2007. The medaka draft genome and insights into vertebrate genome evolution. Nature 447: 714–719. 10.1038/nature05846 [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30: 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B-M, Amores A, Kang S, Ahn D-H, Kim J-H, Kim I-C, Lee JH, Lee SG, Lee H, Lee J, et al. 2019. Antarctic blackfin icefish genome reveals adaptations to extreme environments. Nat Ecol Evol 3: 469–478. 10.1038/s41559-019-0812-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köster J, Rahmann S. 2012. Snakemake—a scalable bioinformatics workflow engine. Bioinformatics 28: 2520–2522. 10.1093/bioinformatics/bts480 [DOI] [PubMed] [Google Scholar]
- Krzywinski MI, Schein JE, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. 2009. Circos: an information aesthetic for comparative genomics. Genome Res 19: 1639–1645. 10.1101/gr.092759.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Suleski M, Hedges SB. 2017. TimeTree: a resource for timelines, timetrees, and divergence times. Mol Biol Evol 34: 1812–1819. 10.1093/molbev/msx116 [DOI] [PubMed] [Google Scholar]
- Li J-T, Wang Q, Huang Yang M-D, Li Q-S, Cui M-S, Dong Z-J, Wang H-W, Yu J-H, Zhao Y-J, Yang C-R, et al. 2021. Parallel subgenome structure and divergent expression evolution of allo-tetraploid common carp and goldfish. Nat Genet 53: 1493–1503. 10.1038/s41588-021-00933-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Wang J, Jaehnig EJ, Shi Z, Zhang B. 2019. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res 47: W199–W205. 10.1093/nar/gkz401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieschke GJ, Currie PD. 2007. Animal models of human disease: zebrafish swim into view. Nat Rev Genet 8: 353–367. 10.1038/nrg2091 [DOI] [PubMed] [Google Scholar]
- Lloyd A, Blary A, Charif D, Charpentier C, Tran J, Balzergue S, Delannoy E, Rigaill G, Jenczewski E. 2018. Homoeologous exchanges cause extensive dosage-dependent gene expression changes in an allopolyploid crop. New Phytol 217: 367–377. 10.1111/nph.14836 [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. 2000. The evolutionary fate and consequences of duplicate genes. Science 290: 1151–1155. 10.1126/science.290.5494.1151 [DOI] [PubMed] [Google Scholar]
- Mandáková T, Lysak MA. 2018. Post-polyploid diploidization and diversification through dysploid changes. Curr Opin Plant Biol 42: 55–65. 10.1016/j.pbi.2018.03.001 [DOI] [PubMed] [Google Scholar]
- Martin KJ, Holland PWH. 2014. Enigmatic orthology relationships between Hox clusters of the African butterfly fish and other teleosts following ancient whole-genome duplication. Mol Biol Evol 31: 2592–2611. 10.1093/molbev/msu202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason AS, Wendel JF. 2020. Homoeologous exchanges, segmental allopolyploidy, and polyploid genome evolution. Front Genet 11: 1014. 10.3389/fgene.2020.01014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama Y, Ito F, Takeda H, Yano T, Okabe M, Kuraku S, Keeley FW, Koshiba-Takeuchi K. 2016. Evolution of the fish heart by sub/neofunctionalization of an elastin gene. Nat Commun 7: 10397. 10.1038/ncomms10397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Yasuike M, Mekuchi M, Iwasaki Y, Ojima N, Fujiwara A, Chow S, Saitoh K. 2017. Rhodopsin gene copies in Japanese eel originated in a teleost-specific genome duplication. Zool Lett 3: 18. 10.1186/s40851-017-0079-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatani Y, McLysaght A. 2017. Genomes as documents of evolutionary history: a probabilistic macrosynteny model for the reconstruction of ancestral genomes. Bioinformatics 33: i369–i378. 10.1093/bioinformatics/btx259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen NTT, Vincens P, Dufayard JF, Roest Crollius H, Louis A. 2022. Genomicus in 2022: comparative tools for thousands of genomes and reconstructed ancestors. Nucleic Acids Res 50: D1025–D1031. 10.1093/nar/gkab1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noutahi E, Semeria M, Lafond M, Seguin J, Boussau B, Guéguen L, El-Mabrouk N, Tannier E. 2016. Efficient gene tree correction guided by genome evolution. PLoS One 11: e0159559. 10.1371/journal.pone.0159559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S, Wolf U, Atkin NB. 1968. Evolution from fish to mammals by gene duplication. Hereditas 59: 169–187. 10.1111/j.1601-5223.1968.tb02169.x [DOI] [PubMed] [Google Scholar]
- Parey E, Louis A, Cabau C, Guiguen Y, Roest Crollius H, Berthelot C. 2020. Synteny-guided resolution of gene trees clarifies the functional impact of whole-genome duplications. Mol Biol Evol 37: 3324–3337. 10.1093/molbev/msaa149 [DOI] [PubMed] [Google Scholar]
- Parey E, Louis A, Montfort J, Bouchez O, Roques C, Iampietro C, Lluch J, Castinel A, Donnadieu C, Desvignes T, et al. 2022. Genome structures resolve the early diversification of teleost fishes. bioRxiv 10.1101/2022.04.07.487469v1 [DOI] [PubMed]
- Pasquier J, Cabau C, Nguyen T, Jouanno E, Severac D, Braasch I, Journot L, Pontarotti P, Klopp C, Postlethwait JH, et al. 2016. Gene evolution and gene expression after whole genome duplication in fish: the PhyloFish database. BMC Genomics 17: 368. 10.1186/s12864-016-2709-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postlethwait JH, Woods IG, Ngo-Hazelett P, Yan YL, Kelly PD, Chu F, Huang H, Hill-Force A, Talbot WS. 2000. Zebrafish comparative genomics and the origins of vertebrate chromosomes. Genome Res 10: 1890–1902. 10.1101/gr.164800 [DOI] [PubMed] [Google Scholar]
- Redmond AK, Gundappa MK, Macqueen DJ, McLysaght A. 2022. Extensive lineage-specific rediploidisation masks shared whole genome duplication in the sturgeon-paddlefish ancestor. bioRxiv 10.1101/2022.05.16.492067v1 [DOI] [PMC free article] [PubMed]
- Rhie A, McCarthy SA, Fedrigo O, Damas J, Formenti G, Koren S, Uliano-Silva M, Chow W, Fungtammasan A, Kim J, et al. 2021. Towards complete and error-free genome assemblies of all vertebrate species. Nature 592: 737–746. 10.1038/s41586-021-03451-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittschof CC, Bukhari SA, Sloofman LG, Troy JM, Caetano-Anollés D, Cash-Ahmed A, Kent M, Lu X, Sanogo YO, Weisner PA, et al. 2014. Neuromolecular responses to social challenge: common mechanisms across mouse, stickleback fish, and honey bee. Proc Natl Acad Sci 111: 17929–17934. 10.1073/pnas.1420369111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson FM, Gundappa MK, Grammes F, Hvidsten TR, Redmond AK, Lien S, Martin SAM, Holland PWH, Sandve SR, Macqueen DJ. 2017. Lineage-specific rediploidization is a mechanism to explain time-lags between genome duplication and evolutionary diversification. Genome Biol 18: 111. 10.1186/s13059-017-1241-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenfeld C, Blanca J, Gallego V, García-Carpintero V, Herranz-Jusdado JG, Pérez L, Asturiano JF, Cañizares J, Peñaranda DS. 2019. De novo European eel transcriptome provides insights into the evolutionary history of duplicated genes in teleost lineages. PLoS One 14: e0218085. 10.1371/journal.pone.0218085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzicka L, Howe DG, Ramachandran S, Toro S, Van Slyke CE, Bradford YM, Eagle A, Fashena D, Frazer K, Kalita P, et al. 2019. The zebrafish information network: new support for non-coding genes, richer gene ontology annotations and the alliance of genome resources. Nucleic Acids Res 47: D867–D873. 10.1093/nar/gky1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacerdot C, Louis A, Bon C, Berthelot C, Roest Crollius H. 2018. Chromosome evolution at the origin of the ancestral vertebrate genome. Genome Biol 19: 166. 10.1186/s13059-018-1559-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzburger W. 2018. Understanding explosive diversification through cichlid fish genomics. Nat Rev Genet 19: 705–717. 10.1038/s41576-018-0043-9 [DOI] [PubMed] [Google Scholar]
- Schartl M. 2014. Beyond the zebrafish: diverse fish species for modeling human disease. Dis Model Mech 7: 181–192. 10.1242/dmm.012245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Session AM, Uno Y, Kwon T, Chapman JA, Toyoda A, Takahashi S, Fukui A, Hikosaka A, Suzuki A, Kondo M, et al. 2016. Genome evolution in the allotetraploid frog Xenopus laevis. Nature 538: 336–343. 10.1038/nature19840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M. 2001. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinforma Oxf Engl 17: 1246–1247. 10.1093/bioinformatics/17.12.1246 [DOI] [PubMed] [Google Scholar]
- Simakov O, Marlétaz F, Yue J-X, O'Connell B, Jenkins J, Brandt A, Calef R, Tung C-H, Huang T-K, Schmutz J, et al. 2020. Deeply conserved synteny resolves early events in vertebrate evolution. Nat Ecol Evol 4: 820–830. 10.1038/s41559-020-1156-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smedley D, Haider S, Ballester B, Holland R, London D, Thorisson G, Kasprzyk A. 2009. Biomart – biological queries made easy. BMC Genomics 10: 22. 10.1186/1471-2164-10-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Som A. 2015. Causes, consequences and solutions of phylogenetic incongruence. Brief Bioinform 16: 536–548. 10.1093/bib/bbu015 [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinforma Oxf Engl 30: 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins GL. 1947. Types of polyploids: their classification and significance. In Advances in genetics (ed. Demerec M), Vol. 1, pp. 403–429. Elsevier, Amsterdam. 10.1016/S0065-2660(08)60490-3 [DOI] [PubMed] [Google Scholar]
- Streisinger G, Walker C, Dower N, Knauber D, Singer F. 1981. Production of clones of homozygous diploid zebra fish (Brachydanio rerio). Nature 291: 293–296. 10.1038/291293a0 [DOI] [PubMed] [Google Scholar]
- Sun H, Wu S, Zhang G, Jiao C, Guo S, Ren Y, Zhang J, Zhang H, Gong G, Jia Z, et al. 2017. Karyotype stability and unbiased fractionation in the paleo-allotetraploid Cucurbita genomes. Mol Plant 10: 1293–1306. 10.1016/j.molp.2017.09.003 [DOI] [PubMed] [Google Scholar]
- Taylor JS, Braasch I, Frickey T, Meyer A, de Peer YV. 2003. Genome duplication, a trait shared by 22,000 species of Ray-Finned fish. Genome Res 13: 382–390. 10.1101/gr.640303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilella AJ, Severin J, Ureta-Vidal A, Heng L, Durbin R, Birney E. 2009. EnsemblCompara GeneTrees: complete, duplication-aware phylogenetic trees in vertebrates. Genome Res 19: 327–335. 10.1101/gr.073585.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace IM, O'Sullivan O, Higgins DG, Notredame C. 2006. M-Coffee: combining multiple sequence alignment methods with T-Coffee. Nucleic Acids Res 34: 1692–1699. 10.1093/nar/gkl091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheim JO, Murrell B, Smith MD, Kosakovsky Pond SL, Scheffler K. 2015. RELAX: detecting relaxed selection in a phylogenetic framework. Mol Biol Evol 32: 820–832. 10.1093/molbev/msu400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittbrodt J, Shima A, Schartl M. 2002. Medaka—a model organism from the far east. Nat Rev Genet 3: 53–64. 10.1038/nrg704 [DOI] [PubMed] [Google Scholar]
- Xie KT, Wang G, Thompson AC, Wucherpfennig JI, Reimchen TE, MacColl ADC, Schluter D, Bell MA, Vasquez KM, Kingsley DM. 2019. DNA fragility in the parallel evolution of pelvic reduction in stickleback fish. Science 363: 81–84. 10.1126/science.aan1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Xu J, Liu G, Chen L, Zhou Z, Peng W, Jiang Y, Zhao Z, Jia Z, Sun Y, et al. 2019. The allotetraploid origin and asymmetrical genome evolution of the common carp Cyprinus carpio. Nat Commun 10: 4625. 10.1038/s41467-019-12644-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakon HH, Lu Y, Zwickl DJ, Hillis DM. 2006. Sodium channel genes and the evolution of diversity in communication signals of electric fishes: convergent molecular evolution. Proc Natl Acad Sci 103: 3675–3680. 10.1073/pnas.0600160103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Zhang B, Lisch D, Ma J. 2017. Patterns and consequences of subgenome differentiation provide insights into the nature of paleopolyploidy in plants. Plant Cell 29: 2974–2994. 10.1105/tpc.17.00595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwaenepoel A, Van de Peer Y. 2019. Inference of ancient whole-genome duplications and the evolution of gene duplication and loss rates. Mol Biol Evol 36: 1384–1404. 10.1093/molbev/msz088 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.