Abstract
Diminazene aceturate (DIZE), an antiparasitic, is an ACE2 activator, and studies show that activators of this enzyme may be beneficial for COVID-19, disease caused by SARS-CoV-2. Thus, the objective was to evaluate the in silico and in vitro affinity of diminazene aceturate against molecular targets of SARS-CoV-2. 3D structures from DIZE and the proteases from SARS-CoV-2, obtained through the Protein Data Bank and Drug Database (Drubank), and processed in computer programs like AutodockTools, LigPlot, Pymol for molecular docking and visualization and GROMACS was used to perform molecular dynamics. The results demonstrate that DIZE could interact with all tested targets, and the best binding energies were obtained from the interaction of Protein S (closed conformation −7.87 kcal/mol) and Mpro (−6.23 kcal/mol), indicating that it can act both by preventing entry and viral replication. The results of molecular dynamics demonstrate that DIZE was able to promote a change in stability at the cleavage sites between S1 and S2, which could prevent binding to ACE2 and fusion with the membrane. In addition, in vitro tests confirm the in silico results showing that DIZE could inhibit the binding between the spike receptor-binding domain protein and ACE2, which could promote a reduction in the virus infection. However, tests in other experimental models with in vivo approaches are needed.
Keywords: Diminazene aceturate, SARS-CoV-2, COVID-19
Graphical abstract
Abbreviations
- ADT
AutoDock Tools
- ATB
Automated Topology Builder
- LS
Local search
- RAS
Renin-Angiotensin System
- RBD
Receptor binding domain
- RMSF
Root mean square fluctuation
- RMSD
Root Mean Square Deviation
1. Introduction
The virus causing COVID-19, SARS-CoV-2; also named as Severe Acute Respiratory Syndrome virus - SARS, was initially identified in China as an enveloped positive-sense RNA virus capable of infecting human cells through binding of the spike protein to Angiotensin-Converting Enzyme II (ACE2) and other factors that aid virus entry [[1], [2], [3]].
The binding of Spike protein to ACE2, an enzyme that participates in the Renin-Angiotensin System (RAS) and has important physiological functions, such as regulation of water and electrolyte balance and blood pressure, can generate harmful effects [4]. Because ACE2 regulates the harmful effects of Ang II, cleaving it into Ang1-7, which binds to MAS receptors generating vasodilation, where the reduction in ACE2 caused by SARS-CoV-2 can lead to pro-inflammatory mechanisms and cardiovascular injury [[5], [6], [7]].
SARS-CoV-2 commonly causes lung cell injury because this virus can encode various proteins that allow it to escape the immune system of the host. In addition, these proteins attack and overactivate more inflammatory and immune cells, inducing a cytokine storm, resulting in severe damage to infected tissues [8].
The critical stage during viral internalization process occurs when receptor binding domain (RBD) binds to ACE2. RBD is found on the virus surfacein the Spike glycoprotein and has two subunits, S1 and S2. Following RBD binding to ACE2, the spike protein is proteolytically cleaved between the two subunits, facilitating fusion with host cell membrane [9,10]. Additionally, Catepsina L (CatL), a cysteine protease, appears to be involved in the activation of protease S on membranous surface of host cell cells [11,12]. The cleavage of ACE2 is necessary for the fusion of SARS-CoV-2 in the cell membrane, a process performed by domain 17 metallopeptidase (ADAM-17), also known as TNF-α Converting Enzyme (TACE), resulting in the downregulation of ACE2 expression on cell surface generating an imbalance in the RAS. Additionally, an increase in ADAM 17 activity can lead to production of TNF-α and IL-6 and other pro-inflammatory cytokines, leading to the inflammatory process during SARS-CoV-2 infection [13,14]. Once inside cells, SARS-CoV-2 uses Mpro, a type 3C protease important in viral replication and transcription [[15], [16], [17]].
It is important to highlight that while ACE2 downregulation in the cell membrane plays a critical role in the pathology of SARS-CoV-2 infection, as it is associated with cardiovascular, pulmonary, and gastrointestinal tract problems; it can generate organ dysfunction in response to infection [13]. Interruption of Ang II degradation due to downregulation of ACE2 may be an important event in COVID-19, so drugs that activate and upregulate the RAS may have benefits in the pathogenesis of COVID-19 [7,18].
Diminazene Aceturate (DIZE: C14H15N7·2C4H7NO3) is an aromatic diamidine that was developed more than six decades ago as an antiparasitic, is an ACE2 activator in several experimental models, contributing to the protective effect of RAS [6]. DIZE has a very rapid absorption rate and short elimination half-life, being metabolized in the liver and redistributed to peripheral tissues for renal excretion [19].
Activation of ACE2 by DIZE could attenuate pulmonary hypertension; in addition, it re-established the balance of the RAS and reduced the inflammatory cells in the peri-infarct área [20,21]. Furthermore, DIZE can reduce the pro-inflammatory cytokines IL-6, IL-12 and TNF-α [22], thus suggesting that it may produce therapeutic benefits in diseases that alter physiological conditions, especially pulmonary hypertension [23].
Even by considering the ability to upregulate ACE2 such as a complication factor due to this enzyme be the host cell entrance, the use of DIZE in the treatment of patients with COVID-19 seems to be interesting if molecular aspects are taken into account [24]. Moreover, others anti-parasitic agents have demonstrated, specially in the ambit of in vitro approaches, anti-viral potential [25]. Therefore, in this study we investigated the effect of DIZE on SARS-CoV-2 targets using a computational approach, e evaluated whether the drug could interfere with the binding of Spike protein and ACE2 (necessary for the virus entry into the cell) by in vitro methodology, which could reduce virus infection.
2. Material and methods
2.1. Molecular docking
The 3D structure of possible proteins from the new coronavirus (SARS-CoV-2) target were obtained from the Protein Data Bank (PDB) [26] with the code PDB ID: ACE2 (1R42), ADAM17 (2DDF), Cathepsin L (2XU3), RBD (6LZG), closed Protein S (6VXX), open Protein S (6VYB), Mpro (6Y2E) and TMPRSS2 (2OQ5). All docking procedures utilized the Autodock 4.2 package [[27], [28], [29]]. Residues of 3D protein structures were protonated using the online server H++ (http://biophysics.cs.vt.edu/hppdetails.php). Protein and ligands were prepared for docking simulations with AutoDock Tools (ADT) version 1.5.6 [30]. The receptor was considered rigid; each ligand was considered flexible. Gasteiger [31] partial charges were calculated after adding fall hydrogens. Non-polar hydrogen atoms of the protein and ligand were subsequently merged. A cubic box of 60 × 60 × 60 points spacing 0.35 Å between the grid points was generated for whole protein target. The affinity grid centers were defined. Global search Lamarckian genetic algorithm (LGA) [32] and the local search (LS) pseudo-Solis and Wets [33] methods were applied in the docking search. Each ligand was subjected to 100 independent runs of docking simulations [34]. Other docking parameters were set as the default values. The resulting docked conformations were clustered into families according to the RMSD. For a more detailed analysis, the coordinates of the selected complexes were chosen by criterion of lowest docking conformation of the cluster with the lowest energy combined with visual inspection.
2.2. Molecular dynamics (MD)
The MD simulations of the complexes selected after molecular docking (Spike_Dize) were performed using GROMACS 2018.1 software [35,36] following Arcanjo and collaborators [37]. Spike glycoprotein (S glycoprotein) model, code P0DTC2, was used from the SWISS-MODEL server [38]. The model is a trimer (A, B, and C chains), and each chain contains 1.162 amino acid residues. The protonation states of histidine residues of model were determined using the H++ [39]. The topologies of the ligands were generated from online repository Automated Topology Builder (ATB) version 3.0 [40]. To increase sampling, Spike_Dize complex MD simulations were run twice (02) for 10 ns using different starting atomic velocities assuming a Maxwellian distribution. Data generated during the last 4 ns of each simulation system, the period defined as production stage, were used for analysis. A total of 80 snapshots, each taken every 100 ps, were obtained during the production stage. The details of the interactions were calculated with LigPlot++ software [41]. A minimum of 50% contact (total of hydrophobic interactions and hydrogen bonds) in the analyzed frames was defined as a criterion of binding efficacy [42].
2.3. ACE2/SARS-CoV-2 spike inhibitor Screening Assay Kit
We tested DIZE (Sigma Aldrich; St. Louis, MO, USA) to inhibit the binding of SARS-CoV-2 spike protein to ACE2 using the SARS-CoV-2 Spike-ACE2 interaction inhibition Screening Assay Kit (Cayman Chemical Company, Ann Arbor, Michigan, USA), according to the manufacturer's instructions. DIZE was used at the following concentrations 2.68; 8.04, 24.12, 76.6, 217.8, and 653.4 μM dilution in DMSO (0.17) or immunoassay buffer (for Spike inhibitor control), concentrations were from according to a published study on docking and in vitro experiments measuring the ability of DIZE to activate ACE2 [6]. Briefly, all reagents used were prepared following the kit recommendations. The assay uses a 96-well plate with a rabbit antibody labeled with the Spike S1 RBD protein of SARS-CoV2 that binds to a plate pre-coated with mouse and rabbit antibodies. Initially, Immunoassay Buffer C was added to the bottom wells; then, the test samples (DIZE or inhibitor control – item n° 402055) were added to all wells, followed by the addition of ACE2 inhibitor screening reagent (item n° 402057) to all wells except the blanks. Then Spike inhibitor screening reagent (item n° 402056) was added, except for blanks and background wells. The plate was incubated for 60 min at room temperature on the shaker. After incubation, the wells were washed, and the Anti-His-HRP conjugate (item n° 402054) was added to each well, except for the blank wells followed by 30 min incubation; after this time, the TMB substrate solution (item n° 400074) was added to each well; finally, HRP stop solution (item n° 10011355) was added, and the absorbance was measured in a microplate reader (SpectraMax, Molecular Devices, USA) at 450 nm %Inhibition was calculated from: (Absorbancecontrol – Absorbancesample/Absorbancecontrol), following kit instructions. Where Absorbancecontrol is equivalent to the reading of wells where no inhibitor is placed, for the interaction between spike protein and ACE2 to occur (100% initial activity according to Cayman assay brochure).
2.4. Statistical analysis
Data represent the means of the results in triplicate and are expressed as mean ± standard error of the mean. For statistically significant differences, it was considered * p < 0.05, analyzed by ANOVA and Tukey's post hoc, after verifying the normality of the data through the Shapiro-wilk test. From the absorbances, after normalization of data, a non-linear regression was performed to calculate the IC50. GraphPad Prism software 8.0 used for statistical analysis.
3. Results
3.1. Docking analysis
The results concerning molecular docking of the diminazene ligand with the 1R42, 2DDF, 2XU3, 6LZG, 6VXX, 6VYB, 6Y2E, and 2OQ5 proteins are presented in Table 1. The best energies obtained from interaction between diminazene aceturate and proteases were with the closed protein S (6VXX) and Mpro (6YE2), the others being described in descending order. The affinity was observed with binding energy equal to −7.87 kcal/mol, and an inhibition constant of 1.71 μM (Table 1; Fig. 1 ). It was possible to observe the interaction with the site active amino acids Ile973, Leu517 and Ala520 via hydrogen bond in ligands. Furthermore, Mpro (6Y2E) had a molecular affinity with diminazene (Fig. 2 ), with binding energy equal to −6.23 kcal/mol, and an inhibition constant of 26.97 μM. Interaction with the amino acids Phe140, Glu166, His41, His164, Cys44, and Thr25 in active site was observed via a hydrogen bond.
Table 1.
Molecular affinity parameters of diminazene ligand with 1R42, 2DDF, 2XU3, 6LZG, 6VXX, 6VYB, 6Y2E and 2OQ5.
| Complex (Protein-ligand) | ΔGbinda (kcal.mol−1) | Kib | Amino acids that interact through hydrogen bondsc | Amino acids that perform hydrophobic interaction c |
|---|---|---|---|---|
| DIM/Closed Protein S (6VXX) | - 7.87 | 1.71 uM | Ile973and Ala520 | Phe565, Thr393, Cys391, Leu518, Asn544, Leu390, His519, Asp979, Arg983, Leu517 and Ser974 |
| DIM/Open Protein S (6VYB) | - 5.8 | 55.64 uM | Pro463, Asp428, Ser514 and Thr430 | Phe515, Phe429, Pro426, Lys462 and Phe464 |
| DIM/Mpro (6Y2E) | - 6.23 | 26.97 uM | Phe140, Glu166, His41, His164, Cys44 and Thr25 | Cys145, Leu141. His 163, Ser144, Asn142, Met49 and Met165 |
| DIM/TMPRSS2 (2OQ5) | - 6.08 | 35.17 uM | Arg41, Ser195 and Asp189 | Gln192, His57, Ser214, Trp215, Glu218, Gly216, Gly226, Ala190, Val213, Cys42 and Gly193 |
| DIM/ADAM17 (2DDF) | - 6.0 | 39.76 uM | Gly362, Gly412, Glu468, Pro366, Cys365, Thr461 and Ser457 | Lys465, Lys460, Gly354, Val364 and Lys367 |
| DIM/RBD (6LZG) | - 5.92 | 45.54 uM | Arg393, Glu402, Glu375, His374 and Ala348 | Tyr385, His401, His378, Pro346, Thr347, Asp382, Asp350 and Asn394 |
| DIM/Cathepsin L (2XU3) | −5.14 | 171.15 uM | Pro59, Gly58, Asn62, Glu63 and Glu95 | Asn66 |
| DIM/ECA2 (1R42) | - 4.06 | 1.05 mM | Glu402, Tyr510, Glu406 and Tyr515 | Arg518, His374, His505 and Phe504 |
Power bond in best conformation.
Best conformation inhibition constant.
Obtained with Ligplot program.
Fig. 1.
3D molecular docking of the protein–ligand complex with 6VXX protein and ligand diminazene illustrating the active binding site (A) with the respective hydrogen bridge interactions (B and C).
Fig. 2.
3D molecular docking of the protein–ligand complex with 6Y2E protein and ligand diminazene illustrating the active binding site (A) with the respective hydrogen bridge interactions (B and C).
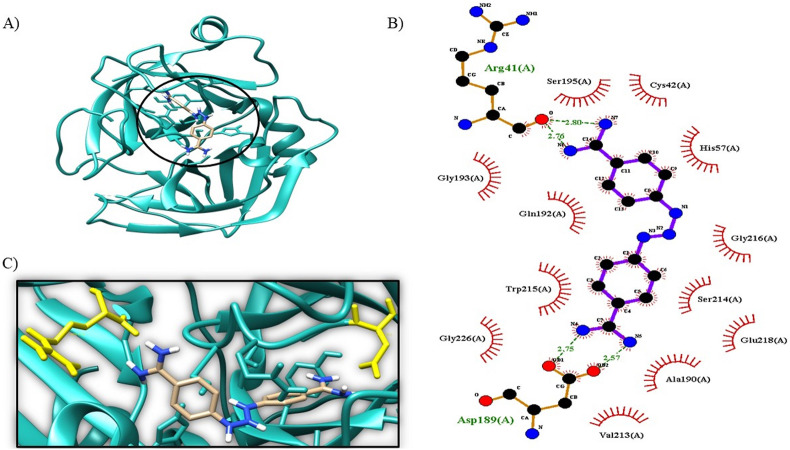
The other proteases and energies obtained from the interaction with diminazene aceturate are described in the table below: open S protein −5,8 kcal/mol and interacting with Pro463, Asp428 (Fig. 3 ), TMPRSS2 connecting to Arg41, Ser195, and Asp189 with the energy of −6,08 kcal/mol (Fig. 4 ), ADAM17–6.0 kcal/mol Gly362, Gly412 and Ser457 (Fig. 5 ), RBD -5.92 kcal/mol with Arg393, Glu402, Glu375 and Ala348 (Fig. 6 ), Cathepsin L −5.14 kcal/mol with Pro59, Gly58, Asn62 and Glu95 (Fig. 7 ) and ACE2 -4,06 kcal/mol Glu402, Tyr510 and Tyr515 (Fig. 8 ).
Fig. 3.
3D molecular docking of the protein–ligand complex with 6VYB protein and ligand diminazene illustrating the active binding site (A) with the respective hydrogen bridge interactions (B and C).
Fig. 4.
3D molecular docking of the protein–ligand complex with 2OQ5 protein and ligand diminazene illustrating the active binding site (A) with the respective hydrogen bridge interactions (B and C).
Fig. 5.
3D molecular docking of the protein–ligand complex with 2DDF protein and ligand diminazene illustrating the active binding site (A) with the respective hydrogen bridge interactions (B and C).
Fig. 6.
3D molecular docking of the protein–ligand complex with 6LZG protein and ligand diminazene illustrating the active binding site (A) with the respective hydrogen bridge interactions (B and C).
Fig. 7.
3D molecular docking of the protein–ligand complex with 2XU3 protein and ligand diminazene illustrating the active binding site (A) with the respective hydrogen bridge interactions (B and C).
Fig. 8.
3D molecular docking of the protein–ligand complex with 1R42 protein and ligand diminazene illustrating the active binding site (A) with the respective hydrogen bridge interactions (B and C).
3.2. Molecular dynamics
Fig. 9 shows that DIZE interacts with the A and B chains of the spike model. DIZE exhibits hydrophobic and hydrogen bond interactions with four residues of the A chain (Tyr396, Phe515, Glu516, and Ser514). In the B chain, DIZE shows hydrophobic interactions with five residues (Tyr200 - 95%, Ile197 - 71%, Lys202 - 68%, Asp228 - 56% and Asp198 - 37%) and hydrogen bonds with four residues (Lys202 - 15%, Asp198 - 15%, Asp228 - 15% and Tyr200 - 2%) (Fig. 9).
Fig. 9.
Interactions for the Spike_DIZE complex. Frequency of hydrophobic contacts – (blue) chain A and (green) chain B. Frequency of hydrogen bonds - (orange) chain A and (red) chain B. The frequencies correspond to the end four of molecular dynamics simulations.
Table 2 shows the Root Mean Aquare Fluctuation (RMSF) values for the residues that interacted with DIZE ligands (Fig. 9). It is observed that there was no significant difference in flexibility in the region of these residues. Fig. 10 shows greater flexibility for residues 682, 685, and 1161 (Fig. 10).
Table 2.
RMSF values for residues that showed interactions with ligands. Root Mean Square Fluctuation values for the residues that interacted with Dize ligands and chain A and B.
| Ligand_chain | Residue | RMSF (nm) |
|---|---|---|
| Dize_A | Tyr396 | 0.1163 |
| The515 | 0.1183 | |
| Glu516 | 0.1499 | |
| Ser514 | 0.1024 | |
| Dize_B | Tyr200 | 0.1198 |
| Ile197 | 0.1187 | |
| Lys202 | 0.0928 | |
| Asp228 | 0.1106 | |
| Asp198 | 0.1637 |
Fig. 10.
RMSF plots for chains A, B and C for ligand DIZE. Root mean square fluctuation (RMSF) - Complex Spike_Dize. Chain A (black), Chain B (red), Chain C (green). Graphical images were produced by XMGRACE software (Turner, 2005).
3.3. In vitro
We examined the ability of DIZE to inhibit S-RBD protein binding to ACE2 in tools 2.68, 8.04, 24.12, 72.6, 217.8, and 653.4 μM. As shown below, the results demonstrate that inhibition rates ranging from 3.82 ± 1.18 to 89.87 ± 7.19%. The highest inhibition rate was achieved at the two highest concentrations tested, 217.8 and 653.4 μM, about 80% (Fig. 11 ). After the analyses, IC50 was equal to 20.62 μM (95% CI = 10.37–37.82 μM).
Fig. 11.
ACE2 S-RBD protein inhibitor screening assay. %inhibition of SARS-Cov-2 spike protein binding to ACE2. Data represent the means of the results in triplicate and are expressed as mean ± standard error of the means. *p < 0.05 in relation to Spike inhibitor control (black bar), followed by ANOVA and Tukey's test.
4. Discussion
COVID-19 stands out as a disease of great concern to public health worldwide [1], having some phases, including the incubation period (up to 5 days), the onset of symptoms (6–7 days), painful breathing (8 days), respiratory distress syndrome (9 days) and severe cases - ICU (10 days) [43]. Since the beginning, several advances have been made to understand the pharmacological targets and therapeutic managements better; however, the suitable search for existing drugs seems valuable [44,45].
An alternative would be drug repositioning, as it is a new strategy for the treatment of emerging diseases. Time becomes crucial in discovering a therapeutic option, where the search for drugs already available on the market and that have new functions could reduce the time, the cost of developing and manufacturing a new therapy, benefiting from detailed information on pharmacology and toxicity which allows for faster trials and clinical use [46,47].
DIZE, an ACE2 activator, has been described in several experimental models showing beneficial effects of RAS [23]. Despite being a veterinary drug, studies have shown that DIZE was used for the treatment of African trypanosomiasis in humans with positive results and minimal adverse effects [48,49], which could justify its use as a new therapeutic option in humans.
Studies have shown that drugs that upregulate ACE2 could be harmful to the patient by stimulating the entry of the virus into cells and the severity of the disease [50,51]. Therefore, Pang; Cui; Zhu [52] do not recommend using DIZE for the treatment of COVID-19. Nonetheless, Nicolau and collaborators [7] propose that DIZE can be used as a new therapeutic strategy in the late stage of pulmonary complications of COVID-19 and that the reestablishment of ACE2 levels could restore homeostasis.
In addition, according to Rodríguez-puertas [53], the treatment of patients using ACE2 activators could bring two therapeutic benefits: i) preventing the binding of the spike protein of SARS-CoV-2 to ACE2 and ii) enabling protective effects of the enzyme in several organs, preventing fibrosis and lung injury. Based on that, we focused our interest on a drug already available on the market and on proteases associated with the life cycle of SARS-CoV-2, because during the cycle, several processes occur that can be used as molecular targets for therapeutic solutions [54]. This study demonstrated for the first time that Diminazene Aceturate can interact with all of the SARS-CoV-2 proteases tested through molecular docking (Table 1). Our results corroborate with Matsoukas and collaborators [55]. These authors showed that, in silico docking and ligand interaction studies, DIZE can interact with some spike protein/ACE2 complex [55].
One of our tested targets was the spike protein of SARS-CoV-2 used in the closed conformation (6VXX) and in the open conformation (6VYB), thus showing that DIZE can bind to proteins regardless of their conformation; however, it demonstrated greater affinity with the closed protein S (−7.87 kcal/mol), thus suggesting that it may act even before SARS-CoV-2 binds to ACE2 and is cleaved by TMPRSS2. As described in the literature, the energies of lower bonds indicate a greater attraction of the ligand for the target thus the ligand with higher affinity can be used as a drug with greater capacity for clinical trials [56,57].
Another target tested was Mpro (6Y2E), a very important cysteine protease for viral multiplication, which makes it a target in an attempt to interrupt the coronavirus life cycle [58,59]. The results suggest that the binding energy found (−6.23 kcal/mol) can block viral replication and prevent the continuation of the virus life cycle. The active site of Mpro is located on the amino acid His41/Cys145 of the catalytic dyad, and Glu166 in molecule A is important to keep the enzyme in its active conformation [12,17]. Our results showed that DIZE acts on the respective amino acids of the active site and that the sensitivity of Mpro to structural disturbances is located around the catalytic site Cys145 [60]. Thus, we propose that DIZE can present satisfactory results in inhibiting this protease, interrupting the proliferation of infectious viral particles.
TMPRSS2, a transmembrane serine protease that also participates in the SARS-CoV-2 infection cycle, was tested, and the results obtained demonstrate that DIZE can interact with the protease (−6.08 kcal/mol); according to the report of Chikhale and collaborators [61], camostat mesylate, a TMPRSS2 inhibitor, obtained energy of −6.23 kcal/mol and according to Gyebi and collaborators [62], it acts on the following amino acids: Gln 192, Arg 41, Ser 195, Trp 215, Ala 190, and Asp 189; however, DIZE had similar energy to the inhibitor and acts on the same and other amino acids: Arg41, Ser195, Asp189 and Gln192, His57, Ser214, Trp215, Glu218, Gly216, Gly226, Ala190, Val213, Cys42, and Gly193. Camostat mesylate could inhibit SARS-CoV-2 infection in human lung cells [63]. It is then suggested that DIZE can also inhibit it, as it has binding energy similar to the inhibitor and acts on the same amino acids; thus, such action would prevent the virus from internalizing.
Cleavage of ACE2 by ADAM 17(2DDF) generates downregulation of ACE2, so its inhibition could exhibit beneficial effects in patients with COVID-19 [64,65]. Furthermore, the energy obtained from the interaction of DIZE with ADAM 17 (−6.0 kcal/mol) demonstrates the possibility of promoting its inhibition. Murumkar and collaborators [66] report that Gly349, Leu 348, and Glu406 are essential active site amino acids and Borah and collaborators [67] demonstrated that IK682, a selective ADAM inhibitor 17, acts on the same amino acids mentioned; however, DIZE was unable to act on any of the amino acids, thus suggesting that the affinity mechanism may occur otherwise and should be investigated.
The RBD (6LZG) represents an important step for SARS-CoV-2 infection, as it is found in protein S and binds to ACE2 present on the cell surface, allowing viral entry and replication [68]. Our results indicate that DIZE can interact with RBD (−5.92 kcal/mol). It was recently identified that amino acid Met82 from ACE2 can bind to Leu472 from RBD and residue Thr487 in RBD interacts with Tyr41 and Lys353 from ACE2 [69], but our results demonstrate that DIZE does not interact with any of these residues; however, such energy obtained suggests that our compound can prevent the virus from binding to ACE2. A recent study argue that the amphoteric nature of DIZE may allow binding to acidic and basic sites of SARS-CoV-2 and possibly block the entry of the virus to ACE2 in the lungs [55].
Cathepsin L (2XU3) plays an important role in SARS-CoV-2 infection, promoting cleavage of the spike glycoprotein S1 subunit [11]. Our results demonstrate that DIZE can interact with CatL (−5.14 kcal/mol). Such results are similar to those found by Vivek-Ananth and collaborators [70], showing that E64-d, an inhibitor of cathepsin proteases including L, showed lower energy (−5.0 kcal/mol) than DIZE, and the catalytic site residues are Cys25, His163, Asp162, Met161, Ala135, Met70, Leu 69, and Gly68; Hardegger and collaborators [71] demonstrate that the amino acid Gly61 from the S3 pocket is found in the active site of the enzyme; however, DIZE does not act directly on the active site amino acids, indicating that the interaction can happen by another mechanism and that the energy generated by our compound is superior to the inhibitor, thus suggesting that it may promote inhibition.
We also demonstrate the in silico relationship of DIZE with the ACE2 (1R42) enzyme that functions as a gateway for the coronavirus in human cells [72]; since this drug is an activator of this enzyme in the RAS [6]. The analysis showed that DIZE promoted interaction with ACE2 (−4.06 kcal/mol). According to Pal and Talukdar [73], the amino acids of the active site of ACE2 would be: Pro426, Ile439, Leu440 and Glu442, and Kulemina and Ostrov [6] demonstrated that DIZE binds outside of the active site and can modulate the catalytic activity of ACE2 in vitro, increasing the maximum initial velocity of the enzyme by 2-fold. The activation of ACE2 by DIZE most likely involves a binding site different from the binding site for AngII and is probably not specific [55].
Our results are consistent with the literature, and we also demonstrate that DIZE does not act directly on the active site of the enzyme; we therefore suggest that its activation occurs by another mechanism and that the interaction with ACE2 may block SARS-CoV-2 binding and prevent its internalization, in addition to avoiding the downregulation of ACE2 and its deleterious effects on the RAS, because studies demonstrate that the activation and recruitment of ACE2 has therapeutic effects, preventing fibrosis and lung injury [53,74]. In addition, DIZE has reduced several pro-inflammatory cytokines, including IL-6, one of those responsible for the seriousness of COVID-19 [22,75]; therefore, we assume that this compound can improve the prognosis of patients with the disease.
Considering that one of the best binding energies obtained from molecular docking was the DIZE/S Protein complex (6VXX) in the closed conformation, molecular dynamics was performed to observe its behavior in the system. Our results demonstrate that DIZE presented an increase in the variation (0.2–0.7 nm) of the RMSF having greater flexibility between residues 682, 685, and 1161, thus suggesting that DIZE can change the stability and flexibility of the Spike protein, causing a change in the protein's structure. The Root Mean Square Deviation (RMSD) measures the distance between the atoms of the structure, in addition to evaluating the stability and displacement of the system [76]. The results found demonstrate a variation of the stability and displacement of the structure, stabilizing at 0.5 nm, indicating that the structure undergoes some conformational change. The RMSD graph for the Spike_dize complex has been added to the Support Information.
Ou and collaborators [77] demonstrate that SARS-CoV-2 has a furin cleavage site between S1 and S2 at residues 682–685 that are important for virus tropism and transmissibility; Egieyeh and collaborators [78] show that high RMSF values indicate greater mobility and less stability in the protein structure, indicating that the coupled drug can prevent the stable interaction between the S protein and the ACE2 enzyme. It is important to highlight that this change in the stability and conformation of the protein between S1 and S2 can prevent protein S cleavage and the externalization of S2 so that fusion with the cell membrane occurs, thus reducing the infection. Therefore, we hypothesize that DIZE can promote the reduction of infection by SARS-CoV-2.
In addition, an in vitro experiment was also carried out to confirm our in silico results. The results obtained demonstrate that DIZE at the concentration of 217.8 and 653.4 μM was able to inhibit the binding of thespike protein RBD to ACE2 in about 80%, and we believe that these results are because the binding of DIZE to S-RBD protein residues reduces the interaction with ACE2, corroborating with the results of the molecular docking. Recently, it was demonstrated using the same experimental approach that the sulfated polysaccharides, polyunsaturated fatty acids, natural and semi-synthetic bile acids, inorganic polyphosphate and stapled peptides [[79], [80], [81], [82], [83]] showed an inhibitory effect similar to those found in this study, preventing the interaction of the spike with ACE2.
Furthermore, the IC50 value obtained (20.62 μM) is similar to those found in the literature evaluating the antiviral effect against SARS-CoV-2 of lopinavir with 26 μM [84]. Recently it was demonstrated that DIZE was able to inhibit proteases, furin and TMPRSS2, in cell cultures, with IC50 of 1.35 and 13.2 μM, respectively [85]. Still, when the concentrations of our compound are increased, its effect on inhibition becomes greater; thus, we suggest that DIZE may have promising effects on the interaction between spike-ACE2 and other proteases, reducing SARS-CoV-2 infection and may generate encouraging clinical effects.
Furthermore, the use of DIZE for humans is very tolerable, one study showed that patients used DIZE for the treatment of human trypanosomiasis [86,87], being regarded as a next-generation drug discovery for the treatment of cardiovascular disease, hypertension and capable of restoring ACE2 functions and promoting cardiovascular and pulmonary protection against SARS-CoV-2, improving the prognosis of patients with COVID-19 [88,89], which can accelerate its use as a therapeutic option in patients with COVID-19.
5. Conclusion
The results found in this article demonstrate that Diminazene Aceturate can interact with all targets tested in molecular docking, acting mainly on the active sites of proteases, and may promote blocking during the viral infection process. Furthermore, molecular dynamics results showed that DIZE can promote a change in amino acid stability at the S protein cleavage site, preventing binding with ACE2. Such results were confirmed through the in vitro test where there was a reduction in the binding rate between the S-RBD protein and ACE2. Therefore, considering the results found and the pharmacological activities described in the literature, we hypothesized that DIZE could interact with SARS-CoV-2 molecular targets related to viral entry and replication, preventing viral infection, reducing the inflammatory process, however, other experimental models with in vivo approaches are needed to confirm this hypothesis.
Funding
None.
Author statement
Esley da S. Santos: Conceptualization, investigation, methodology, research, writing and editing, writing - original draft, visualization. Priscila C. Silva, Luiz F. L. S. Teixeira: Investigation, formal analysis. Paulo Sergio de A. Sousa: Programs, methodology, supervision, validation, investigation. Gabriella Pacheco: Formal analysis, data curation, writing, proofreading, editing. Aline R. de Araujo: Formal analysis, data curation. Francisca B. M. Sousa: writing, proofreading, editing. Romulo O. Barros: Programs, methodology, formal analysis. Ricardo M. Ramos: Programs, methodology, formal analysis. Jefferson A. Rocha: Programs, methodology, validation. Lucas A. D. Nicolau: Conceptualization, data curation, writing, proofreading, editing. Jand V. R. Medeiros: Writing - Original draft, proofreading and editing; Conceptualization; Supervision; Project administration; Acquisition of financing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
The authors are grateful for the financial support of the National Council for Scientific and Technological Development–CNPq (Brazil) and the Foundation for Research Support of the State of Piauí–FAPEPI.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cbi.2022.110161.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Fig. S1.
RMSD plots for chains A, B and C for ligand Dize. Root Mean Square Deviation – RMSD.
Data availability
Data will be made available on request.
References
- 1.Rothan H.A., Byrareddy S.N. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J. Autoimmun. 2020 doi: 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.OF THE INTERNATIONAL, Coronaviridae Study Group et al. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5(4):536. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vaduganathan M., Vardeny O., Michel T. Original: renin–angiotensin–aldosterone system inhibitors in patients with covid. Cell. 2020 doi: 10.1056/NEJMsr2005760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perlot T., Penninger J.M. ACE2–From the renin–angiotensin system to gut microbiota and malnutrition. Microb. Infect. 2013;15(13):866–873. doi: 10.1016/j.micinf.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kulemina L.V., Ostrov D.A. Prediction of off-target effects on angiotensin-converting enzyme 2. J. Biomol. Screen. 2011;16(8):878–885. doi: 10.1177/1087057111413919. [DOI] [PubMed] [Google Scholar]
- 7.Nicolau L.A.D., Nolêto I.R.S.G., Medeiros J.V.R. Could a specific ACE2 activator drug improve the clinical outcome of SARS-CoV-2? A potential pharmacological insight. Expet Rev. Clin. Pharmacol. 2020;13(8):807–811. doi: 10.1080/17512433.2020.1798760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu Z., Lian X., Su X., Wu W., Marraro G.A., Zeng Y. From SARS and MERS to COVID-19: a brief summary and comparison of severe acute respiratory infections caused by three highly pathogenic human coronaviruses. Respir. Res. 2020;21(1):1–14. doi: 10.1186/s12931-020-01479-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burgueño J.F., Reich A., Hazime H., Quintero M.A., Fernandez I., Fritsch J., Santander A.M., Brito N., Damas O.M., Deshpande A., Kerman D.H., Zhang L., Gao Z., Ban Y., Wang L., Pignac-Kobinger J., Abreu M.T. Expression of SARS-CoV 2 entry molecules ACE2 and TMPRSS2 in the gut of patients with IBD. Inflamm. Bowel Dis. 2020;26(6):797–808. doi: 10.1093/ibd/izaa085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sorokina M., Teixeira J.M., Barrera-Vilarmau S., Paschke R., Papasotiriou I., Rodrigues J.P., Kastritis P.L. Structural models of human ACE2 variants with SARS-CoV 2 Spike protein for structure-based drug design. Sci. Data. 2020;7(1):1–10. doi: 10.6084/m9.figshare.12902498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu T., Luo S., Libby P., Shi G.P. Cathepsin L-selective inhibitors: a potentially promising treatment for COVID-19 patients. Pharmacol. Therapeut. 2020;213 doi: 10.1016/j.pharmthera.2020.107587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang X., Zhou Y., Yu X., Huang Q., Fang W., Li J., Bonventre J.V., Sukhova G.K., Libby P., Shi G.P. Differential roles of cysteinyl cathepsins in TGF-β signaling and tissue fibrosis. iScience. 2019;19:607–622. doi: 10.1016/j.isci.2019.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gheblawi M., Wang K., Viveiros A., Nguyen Q., Zhong J.C., Turner A.J., Raizada M.K., Grant M.B., Oudit G.Y. Angiotensin-converting enzyme 2: SARS-CoV 2 receptor and regulator of the renin-angiotensin system: celebrating the 20th anniversary of the discovery of ACE2. Circ. Res. 2020;126(10):1456–1474. doi: 10.1161/CIRCRESAHA.120.317015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schumacher N., Rose-John S. Adam17 activity and IL-6 trans-signaling in inflammation and cancer. Cancers. 2019;11(11):1736. doi: 10.3390/cancers11111736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hegyi A., Ziebuhr J. Conservation of substrate specificities among coronavirus main proteases. J. Gen. Virol. 2002;83(3):595–599. doi: 10.1099/0022-1317-83-3-595. [DOI] [PubMed] [Google Scholar]
- 16.Rut W., Groborz K., Zhang L., Sun X., Zmudzinski M., Pawlik B., Wang X., Jochmans D., Neyts J., Mlynarski W., Hilgenfeld M., Drag M. SARS-CoV-2 M pro inhibitors and activity-based probes for patient-sample imaging. Nat. Chem. Biol. 2021;17(2):222–228. doi: 10.1038/s41589-020-00689-z. [DOI] [PubMed] [Google Scholar]
- 17.Ullrich S., Nitsche C. The SARS-CoV 2 main protease as drug target. Bioorg. Med. Chem. Lett. 2020 doi: 10.1016/j.bmcl.2020.127377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nicolau L.A.D., Magalhães P.J.C., Vale M.L. What would Sérgio Ferreira say to your physician in this war against COVID-19: how about kallikrein/kinin system? Med. Hypotheses. 2020;143 doi: 10.1016/j.mehy.2020.109886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller D.M., Swan G.E., Lobetti R.G., Jacobson L.S. The pharmacokinetics of diminazene ceturate after intramuscular administration in healthy dogs. J. South Afr. Vet. Assoc. 2005;76(3):146–150. doi: 10.4102/jsava.v76i3.416. [DOI] [PubMed] [Google Scholar]
- 20.Qi Y., Zhang J., Cole-Jeffrey C.T., Shenoy V., Espejo A., Hanna M., Chunjuan C., Pepine C.J., Katovich M.J., Raizada M.K. Diminazene aceturate enhances angiotensin-converting enzyme 2 activity and attenuates ischemia-induced cardiac pathophysiology. Hypertension. 2013;62(4):746–752. doi: 10.1161/HYPERTENSIONAHA.113.01337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shenoy V., Gjymishka A., Jarajapu Y.P., Qi Y., Afzal A., Rigatto K., Ferreira A.J., Fraga-Silva R.A., Kearns P., Douglas J.Y., Agarwal D., Mubarak K.K., Bradford C., Kennedy W.R., Joo Y., Rathinasabapathy A., Bruce E., Gupta D., Cardounel A.J., Mocco J., Patel J.M., Francis J., Grant M.B., Katovich M.J., Raizada M.K. Diminazene attenuates pulmonary hypertension and improves angiogenic progenitor cell functions in experimental models. Am. J. Respir. Crit. Care Med. 2013;187(6):648–657. doi: 10.1164/rccm.201205-0880OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuriakose S., Muleme H.M., Onyilagha C., Singh R., Jia P., Uzonna J.E. Diminazene aceturate (Berenil) modulates the host cellular and inflammatory responses to Trypanosoma congolense infection. PLoS One. 2012;7(11) doi: 10.1371/journal.pone.0048696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuriakose S., Muleme H., Onyilagha C., Okeke E., Uzonna J.E. Diminazene aceturate (Berenil) modulates LPS induced pro-inflammatory cytokine production by inhibiting phosphorylation of MAPKs and STAT proteins. Innate Immun. 2014;20(7):760–773. doi: 10.1177/1753425913507488. [DOI] [PubMed] [Google Scholar]
- 24.Marilda L.A.D.M., Liliane D.A., Rodrigo A.F.-S., Letícia A.S.P., Heder J.R., Gustavo B.M., Vinayak S., Mohan K.R., Anderson J.F. Anti-hypertensive effects of diminazene aceturate: an angiotensin- converting enzyme 2 activator in rats. Protein Pept. Lett. 2016;23:9–16. doi: 10.2174/0929866522666151013130550. [DOI] [PubMed] [Google Scholar]
- 25.Panahi Y., Dadkhah M., Talei S., Gharari Z., Asghariazar V., Abdolmaleki A., Matin S., Molaei S. Can anti-parasitic drugs help control COVID-19? Future Virol. 2022 May;17(5):315–339. doi: 10.2217/fvl-2021-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The protein data bank. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goodsell D.S. Computational docking of biomolecular complexes with AutoDock. Cold Spring Harb. Protoc. 2009;2009(5) doi: 10.1101/pdb.prot5200. p. pdb. prot5200. 10.1101/pdb.prot5200. [DOI] [PubMed] [Google Scholar]
- 28.Goodsell D.S., Morris G.M., Olson A.J. Automated docking of flexible ligands: applications of AutoDock. J. Mol. Recogn. 1996;9(1):1–5. doi: 10.1002/(SICI)1099-1352(199601)9:1<1::AID-JMR241>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 29.Morris G.M., Huey R., Olson A.J. Using autodock for ligand‐receptor docking. Curr. Protocols Bioinf. 2008;24(1):8. doi: 10.1002/0471250953.bi0814s24. 14. 1-8.14. 40. [DOI] [PubMed] [Google Scholar]
- 30.Sanner M.F. Python: a programming language for software integration and development. J. Mol. Graph. Model. 1999;17(1):57–61. [PubMed] [Google Scholar]
- 31.Gasteiger J., Marsili M. Iterative partial equalization of orbital electronegativity—a rapid access to atomic charges. Tetrahedron. 1980;36(22):3219–3228. doi: 10.1016/0040-4020(80)80168-2. [DOI] [Google Scholar]
- 32.Morris G.M., Goodsell D.S., Halliday R.S., Huey R., Hart W.E., Belew R.K., Olson A.J. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998;19(14):1639–1662. doi: 10.1002/(SICI)1096-987X(19981115)19:14<1639::AID-JCC10>3.0.CO;2-B. [DOI] [Google Scholar]
- 33.Solis F.J., Wets R.J.B. Minimization by random search techniques. Math. Oper. Res. 1981;6(1):19. doi: 10.1287/moor.6.1.19. 3. [DOI] [Google Scholar]
- 34.Rocha J.A., Rego N.C., Carvalho B.T., Silva F.I., Sousa J.A., Ramos R.M., Passos I.N.G., Moraes J., Leite J.R.S.A., Lima F.C. Computational quantum chemistry, molecular docking, and ADMET predictions of imidazole alkaloids of Pilocarpus microphyllus with schistosomicidal properties. PLoS One. 2018;13(6) doi: 10.1371/journal.pone.0198476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oostenbrink C., Villa A., Mark A.E., Van Gunsteren W.F. A biomolecular force field based on the free enthalpy of hydration and solvation: the GROMOS force‐field parameter sets 53A5 and 53A6. J. Comput. Chem. 2004;25(13):1656–1676. doi: 10.1002/jcc.20090. [DOI] [PubMed] [Google Scholar]
- 36.Abraham M.J., Van Der Spoel D., Lindahl E., Hess B. The GROMACS development team. GROMACS user manual version. 2018. www.gromacs.org 1.
- 37.Arcanjo D.D., Vasconcelos A.G., Nascimento L.A., Mafud A.C., Plácido A., Alves M.M., Delerue-Matos C., Bemquerer M.P., Vale N., Gomes P., Oliveira E.B., Lima F.C.A., Mascarenhas Y.P., Carvalho F.A., Simonsen U., Ramos R.M., Leite J.R.S. Structure-function studies of BPP-BrachyNH2 and synthetic analogues thereof with Angiotensin I-Converting Enzyme. Eur. J. Med. Chem. 2017;139:401–411. doi: 10.1016/j.ejmech.2017.08.019. [DOI] [PubMed] [Google Scholar]
- 38.Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., Beer T.A.P., Rempfer C., Bordoli L., Lepore R., Schwede T. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46(W1):W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gordon J.C., Myers J.B., Folta T., Shoja V., Heath L.S., Onufriev A. H++: a server for estimating p K as and adding missing hydrogens to macromolecules. Nucleic Acids Res. 2005;33(suppl_2):W368–W371. doi: 10.1093/nar/gki464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stroet M., Caron B., Visscher K.M., Geerke D.P., Malde A.K., Mark A.E. Automated topology builder version 3.0: prediction of solvation free enthalpies in water and hexane. J. Chem. Theor. Comput. 2018;14(11):5834–5845. doi: 10.1021/acs.jctc.8b00768. [DOI] [PubMed] [Google Scholar]
- 41.Laskowski R.A., Swindells M.B. LigPlot+: multiple ligand–protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011;51(10):2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- 42.Ramos R.M., Perez J.M., Baptista L.A., de Amorim H.L.N. Interaction of wild type, G68R and L125M isoforms of the arylamine-N-acetyltransferase from Mycobacterium tuberculosis with isoniazid: a computational study on a new possible mechanism of resistance. J. Mol. Model. 2012;18(9):4013–4024. doi: 10.1007/s00894-012-1383-6. [DOI] [PubMed] [Google Scholar]
- 43.Chakraborty C., Sharma A.R., Sharma G., Bhattacharya M., Lee S.S. SARS-CoV-2 causing pneumonia-associated respiratory disorder (COVID-19): diagnostic and proposed therapeutic options. Eur. Rev. Med. Pharmacol. Sci. 2020;24(7):4016–4026. doi: 10.26355/eurrev_202004_20871. [DOI] [PubMed] [Google Scholar]
- 44.Alexpandi R., De Mesquita J.F., Pandian S.K., Ravi A.V. Quinolines-based SARS-CoV 2 3CLpro and RdRp inhibitors and Spike-RBD-ACE2 inhibitor for drug-repurposing against COVID-19: na in silico analysis. Front. Microbiol. 2020;11:1796. doi: 10.3389/fmicb.2020.01796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tu Y.F., Chien C.S., Yarmishyn A.A., Lin Y.Y., Luo Y.H., Lin Y.T., Lai W.Y., Yang D.M., Chou S.J., Yang Y.P., Wang M.L., Chiou S.H. A review of SARS-CoV 2 and the ongoing clinical trials. Int. J. Mol. Sci. 2020;21(7):2657. doi: 10.3390/ijms21072657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pushpakom S., Iorio F., Eyers P.A., Escott K.J., Hopper S., Wells A., Doig A., Guilliams T., Latimer J., McNamee C., Norris A., Sanseau P., Cavalla D., Pirmohamed M. Drug repurposing: progress, challenges and recommendations. Nat. Rev. Drug Discov. 2019;18(1):41–58. doi: 10.1038/nrd.2018.168. [DOI] [PubMed] [Google Scholar]
- 47.Ton A.-T., Gentile F., Hsing M., Ban F., Cherkasov A. Rapid identification of potential inhibitors of SARS‐CoV‐2 main protease by deep docking of 1.3 billion compounds. Mol. Inf. 2022;39(8):e2000028. doi: 10.1002/minf.202000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hutchinson M.P., Watson H.J.C. Berenil in the treatment of Trypanosoma gambiense infection in man. Trans. R. Soc. Trop. Med. Hyg. 1962;56(3):227–230. doi: 10.1016/0035-9203(62)90158-x. [DOI] [PubMed] [Google Scholar]
- 49.Abaru D.E., Liwo D.A., Isakina D., Okori E.E. Retrospective long-term study of effects of berenil by follow-up of patients treated since 1965. Tropenmed. Parasitol. 1984;35(3):148–150. [PubMed] [Google Scholar]
- 50.Fang L., Karakiulakis G., Roth M. Are patients with hypertension and diabetes mellitus at increased risk for COVID-19 infection? Lancet Respir. Med. 2020;8(4):e21. doi: 10.1016/S2213-2600(20)30116-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Diaz J.H. Hypothesis: angiotensin-converting enzyme inhibitors and angiotensin receptor blockers may increase the risk of severe. J. Trav. Med. 2020;382:1653–1659. doi: 10.1093/jtm/taaa041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pang X., Cui Y., Zhu Y. Recombinant human ACE2: potential therapeutics of SARS-CoV-2 infection and its complication. Acta Pharmacol. Sin. 2020;41(9):1255–1257. doi: 10.1038/s41401-020-0430-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rodríguez‐Puertas R. ACE2 activators for the treatment of COVID 19 patients. J. Med. Virol. 2020;92(10):1701–1702. doi: 10.1002/jmv.25992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ortega J.T., Serrano M.L., Jastrzebska B. Class AG protein-coupled receptor antagonist famotidine as a therapeutic alternative against SARS-CoV 2: an in silico analysis. Biomolecules. 2020;10(6):954. doi: 10.3390/biom10060954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Matsoukas J.M., Gadanec K.L., Zulli A., Apostolopoulos V., Konstantinos K., Ligielli I., Moschovou K., Georgiou N., Plotas P., Chasapis C.T., Moore G., Ridgway H., Mavromoustakos T. Diminazene aceturate reduces angiotensin II constriction and interacts with the spike protein of severe acute respiratory syndrome coronavirus 2. Biomedicines. 2022;10(7):1731. doi: 10.3390/biomedicines10071731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen D., Oezguen N., Urvil P., Ferguson C., Dann S.M., Savidge T.C. Regulation of protein-ligand binding affinity by hydrogen bond pairing. Sci. Adv. 2016;2(3) doi: 10.1126/sciadv.1501240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Raj S., Sasidharan S., Dubey V.K., Saudagar P. Identification of lead molecules against potential drug target protein MAPK4 from L. donovani: an in-silico approach using docking, molecular dynamics and binding free energy calculation. PLoS One. 2019;14(8) doi: 10.1371/journal.pone.0221331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tomic N., Pojskic L., Kalajdzic A., Ramic J., Kadric N.L., Ikanovic T., Maksimovic M., Pojskic N. Screening of preferential binding affinity of selected natural compounds to SARS-CoV 2 proteins using in silico methods. EJMO. 2020;4:319–323. doi: 10.14744/ejmo.2020.72548. [DOI] [Google Scholar]
- 59.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Xiao G., Rao Z., Yang H. Structure of M pro from SARS-CoV 2 and discovery of its inhibitors. Nature. 2020;582(7811):289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 60.Estrada E. Topological analysis of SARS-CoV 2 main protease. Chaos: Interdiscip. J. Nonlinear Sci. 2020;30(6) doi: 10.1063/5.0013029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chikhale R.V., Gupta V.K., Eldesoky G.E., Wabaidur S.M., Patil S.A., Islam M.A. Identification of potential anti-TMPRSS2 natural products through homology modelling, virtual screening and molecular dynamics simulation studies. J. Biomol. Struct. Dyn. 2020:1–16. doi: 10.1080/07391102.2020.1798813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gyebi G.A., Adegunloye A.P., Ibrahim I.M., Ogunyemi O.M., Afolabi S.O., Ogunro O.B. Prevention of SARS-CoV 2 cell entry: insight from in silico interaction of drug-like alkaloids with spike glycoprotein, human ACE2, and TMPRSS2. J. Biomol. Struct. Dyn. 2020:1–25. doi: 10.1080/07391102.2020.1835726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV 2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Heurich A., Hofmann-Winkler H., Gierer S., Liepold T., Jahn O., Pöhlmann S. TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein. J. Virol. 2014;88(2):1293–1307. doi: 10.1128/JVI.02202-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Palau V., Riera M., Soler M.J. ADAM17 inhibition may exert a protective effect on COVID-19. Nephrol. Dial. Transplant. 2020;35(6):1071–1072. doi: 10.1093/ndt/gfaa093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Murumkar P.R., Sharma M.K., Giridhar R., Yadav M.R. Virtual screening-based identification of lead molecules as selective TACE inhibitors. Med. Chem. Res. 2015;24(1):226–244. doi: 10.1007/s00044-014-1097-7. [DOI] [Google Scholar]
- 67.Borah P.K., Chakraborty S., Jha A.N., Rajkhowa S., Duary R.K. In silico approaches and proportional odds model towards identifying selective adam17 inhibitors from anti-inflammatory natural molecules. J. Mol. Graph. Model. 2016;70:129–139. doi: 10.1016/j.jmgm.2016.10.003. 2016. [DOI] [PubMed] [Google Scholar]
- 68.Braz H.L.B., de Moraes Silveira J.A., Marinho A.D., de Moraes M.E.A., de Moraes Filho M.O., Monteiro H.S.A., Jorge R.J.B. In silico study of azithromycin, chloroquine and hydroxychloroquine and their potential mechanisms of action against SARS-CoV 2 infection. Int. J. Antimicrob. Agents. 2020;56(3) doi: 10.1016/j.ijantimicag.2020.106119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Z., Xiao X., Wei X., Li J., Yang J., Tan H., Zhu J., Zhang Q., Wu J., Liu L. Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARS‐CoV‐2. J. Med. Virol. 2020;92(6):595–601. doi: 10.1002/jmv.25726. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vivek-Ananth R.P., Rana A., Rajan N., Biswal H.S., Samal A. In silico identification of potential natural product inhibitors of human proteases key to SARS-CoV-2 infection. Molecules. 2020;25(17):3822. doi: 10.3390/molecules25173822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hardegger L.A., Kuhn B., Spinnler B., Anselm L., Ecabert R., Stihle M., Gsell B., Thoma R., Diez J., Benz J., Plancher J.M., Hartmann G., Banner D.W., Haap W., Diederich F. Systematic investigation of halogen bonding in protein–ligand interactions. Angew. Chem. Int. Ed. 2011;50(1):314–318. doi: 10.1002/anie.201006781. [DOI] [PubMed] [Google Scholar]
- 72.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292. doi: 10.1016/j.cell.2020.02.058. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pal S., Talukdar A. 2020. Compilation of Potential Protein Targets for SARS-CoV 2: Preparation of Homology Model and Active Site Determination for Future Rational Antiviral Design. [DOI] [Google Scholar]
- 74.Imai Y., Kuba K., Rao S., Huan Y., Guo F., Guan B., Yang P., Sarao R., Wada T., Crackower M.A., Hein L., Jiang C., Penninger J.M. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature. 2005;436(7047):112–116. doi: 10.1038/nature03712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Potere N., Batticciotto A., Vecchié A., Porreca E., Cappelli A., Abbate A., Dentali F., Bonaventura A. The role of IL-6 and IL-6 blockade in COVID-19. Expet Rev. Clin. Immunol. 2021:1–17. doi: 10.1080/1744666X.2021.1919086. [DOI] [PubMed] [Google Scholar]
- 76.Martínez L. Automatic identification of mobile and rigid substructures in molecular dynamics simulations and fractional structural fluctuation analysis. PLoS One. 2015;10(3) doi: 10.1371/journal.pone.0119264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Chen T., Hu J., Xiang Z., Mu Z., Chen X., Wang J., Qian Z. 2020. Characterization of Spike Glycoprotein of 2019-nCoV on Virus Entry and its Immune Cross-Reactivity with Spike Glycoprotein of SARS-CoV. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Egieyeh S., Egieyeh E., Malan S., Christofells A., Fielding B. Computational drug repurposing strategy predicted peptide-based drugs that can potentially inhibit the interaction of SARS-CoV-2 spike protein with its target (humanACE2) PLoS One. 2021;16(1) doi: 10.1371/journal.pone.0245258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kwon P.S., Oh H., Kwon S.J., Jin W., Zhang F., Fraser K., Hong J.J., Linhardt R.J., Dordick J.S. Sulfated polysaccharides effectively inhibit SARS-CoV-2 in vitro. Cell Discov. 2020;6(1):1–4. doi: 10.1038/s41421-020-00192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Goc A., Niedzwiecki A., Rath M. Polyunsaturated ω-3 fatty acids inhibit ACE2-controlled SARS-CoV-2 binding and cellular entry. Sci. Rep. 2021;11(1):1–12. doi: 10.1038/s41598-021-84850-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Carino A., Moraca F., Fiorillo B., Marchianò S., Sepe V., Biagioli M., Finamore C., Bozza S., Francisci D., Distrutti E., Catalanotti B., Zampella A., Fiorucci S. Hijacking SARS-CoV-2/ACE2 receptor interaction by natural and semi-synthetic steroidal agents acting on functional pockets on the receptor binding domain. Front. Chem. 2020;8:846. doi: 10.3389/fchem.2020.572885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Neufurth M., Wang X., Tolba E., Lieberwirth I., Wang S., Schröder H.C., Müller W.E. The inorganic polymer, polyphosphate, blocks binding of SARS-CoV-2 spike protein to ACE2 receptor at physiological concentrations. Biochem. Pharmacol. 2020;182 doi: 10.1016/j.bcp.2020.114215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Maas M.N., Hintzen J.C., Löffler P.M., Mecinović J. Targeting SARS-CoV-2 spike protein by stapled hACE2 peptides. Chem. Commun. 2021;57(26):3283–3286. doi: 10.1039/D0CC08387A. [DOI] [PubMed] [Google Scholar]
- 84.Choy K., Wong A.Y., Kaewpreedee P., Sai S.F., Chen D., Hui K.P.Y., Chu D.K.W., Chan M.C.W., Cheung P. P. Huang, Peiris X., Yen M., Remdesivir H. lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antivir. Res. 2020;178 doi: 10.1016/j.antiviral.2020.104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xu Y.-M., Inácio M.C., Liu M.X., Gutalilaka A.A.L. Discovery of diminazene as a dual inhibitor of SARS-CoV-2 human host proteases TMPRSS2 and furin using cell-based assays. Curr. Res. Chem. Biol. 2022;2 doi: 10.1016/j.crchbi.2022.100023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Oliveira G.L.S., Freitas R.M. Diminazene aceturate—an antiparasitic drug of antiquity: advances in pharmacology & therapeutics. Pharmacol. Res. 2015;102:138–157. doi: 10.1016/j.phrs.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 87.Peregrine A.S., Mamman M. Pharmacology of diminazene: a review. Acta Trop. 1993;54(3–4):185–203. doi: 10.1016/0001-706X(93)90092-P. [DOI] [PubMed] [Google Scholar]
- 88.Gjymishka A., Kulemina L.V., Shenoy V., Katovich M., Ostrov D.A., Raizada M.K. Diminazene aceturate is an ACE2 activator and a novel antihypertensive drug. Faseb. J. 2010;24(3):1032–1033. doi: 10.1096/fasebj.24.1_supplement.1032.3. 1032. [DOI] [Google Scholar]
- 89.Kate gadanec L., Qaradkhi T., McSweeney K.R., Ali B.A., Zulli A., Apostolopoulos V. Dual targeting of toll-like receptor 4 and angiotensin-converting enzyme 2: a proposed approach to SARS-CoV-2 treatment. Future Microbiol. 2021;16(4):205–209. doi: 10.2217/fmb-2021-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.