Significance
Genome sequences are being produced for more and more eukaryotic species. The bulk of these genomes are composed of parasitic, self-mobilizing transposable elements (TEs) that play important roles in organismal evolution. Thus there is a pressing need for developing software that can accurately identify the diverse set of TEs dispersed in genome sequences. Here we introduce RepeatModeler2, an easy-to-use package for the production of reference TE libraries which can be applied to any eukaryotic species. Through several major improvements over the previous version, RepeatModeler2 is able to produce libraries that recapitulate the known composition of three model species with some of the most complex TE landscapes. Thus RepeatModeler2 will greatly enhance the discovery and annotation of TEs in genome sequences.
Keywords: genome annotation, mobile genetic elements, transposon families
Abstract
The accelerating pace of genome sequencing throughout the tree of life is driving the need for improved unsupervised annotation of genome components such as transposable elements (TEs). Because the types and sequences of TEs are highly variable across species, automated TE discovery and annotation are challenging and time-consuming tasks. A critical first step is the de novo identification and accurate compilation of sequence models representing all of the unique TE families dispersed in the genome. Here we introduce RepeatModeler2, a pipeline that greatly facilitates this process. This program brings substantial improvements over the original version of RepeatModeler, one of the most widely used tools for TE discovery. In particular, this version incorporates a module for structural discovery of complete long terminal repeat (LTR) retroelements, which are widespread in eukaryotic genomes but recalcitrant to automated identification because of their size and sequence complexity. We benchmarked RepeatModeler2 on three model species with diverse TE landscapes and high-quality, manually curated TE libraries: Drosophila melanogaster (fruit fly), Danio rerio (zebrafish), and Oryza sativa (rice). In these three species, RepeatModeler2 identified approximately 3 times more consensus sequences matching with >95% sequence identity and sequence coverage to the manually curated sequences than the original RepeatModeler. As expected, the greatest improvement is for LTR retroelements. Thus, RepeatModeler2 represents a valuable addition to the genome annotation toolkit that will enhance the identification and study of TEs in eukaryotic genome sequences. RepeatModeler2 is available as source code or a containerized package under an open license (https://github.com/Dfam-consortium/RepeatModeler, http://www.repeatmasker.org/RepeatModeler/).
Most eukaryotic genomes contain a large number of interspersed repeats that by and large represent copies of transposable elements (TEs) at varying stages of evolutionary decay (1–5). TEs are genomic sequences capable of mobilization and replication, generating complex patterns of repeats that account for up to 85% of eukaryotic genome content (6). Different organisms have diverse TE landscapes, including a wide range of abundances, activity levels, and sequence degradation levels (7, 8). As mutagens and major contributors to the organization, rearrangement, and regulation of the genome, TEs have had a profound impact on organismal evolution (reviewed in ref. 4). Our understanding of the biological impact of TEs has grown steadily through the study of both model and nonmodel organisms from which whole-genome sequences can now be routinely assembled. With each new species sequenced comes the challenge of identifying its unique set of TE families, which remains a tedious and largely manual endeavor. Yet, the accurate identification of TEs and other repeats is a prerequisite to nearly all other genomic analysis, including the annotation of genes (9).
What makes TEs so potent in remodeling the genome but also challenging to annotate is their diversity in structures and sequences, which greatly vary across species (3, 4). There are two major classes of TEs (reviewed in refs. 10–12; https://www.dfam.org/classification): Class I retroelements replicate and transpose via an RNA intermediate; while class II elements (or DNA transposons) are mobilized via a DNA intermediate. Class I elements include long and short interspersed elements (LINEs, SINEs) and long terminal repeat (LTR) retrotransposons. The most common class II transposons are TIR (terminal inverted repeats) elements, which transpose via a “cut-and-paste” mechanism (13). But other class II elements, such as Helitrons, also use replicative mechanisms (14–16). Within each class, TE sequences are extremely diverse and evolve rapidly (4, 10, 17). Additionally, once integrated in the host genome, each element is subject to mutations, such as point mutations, and a vast array of rearrangements, including internal deletions, truncations, and nested insertions. The vast sequence diversity of TEs combined with the complexity of mutations that occur after insertion makes automated TE identification and classification a daunting task.
The most elementary level of classification of TEs is the family, which designates interspersed genomic copies derived from the amplification of an ancestral progenitor sequence (10). Each TE family can be represented by a consensus sequence approximating that of the ancestral progenitor. Such consensus sequence can be recreated from a multiple alignment of individual genomic copies (or “seeds”) from which each ancestral nucleotide can be inferred based on a majority rule along the alignment. Similarly, the seed alignment may be used to generate a profile Hidden Markov Model (HMM) for each family. Consensus TE sequences and HMMs are used for many downstream applications in the study and annotation of genomes. Notably, they are used to annotate or “mask” the genome using RepeatMasker, which is a prerequisite for gene annotation (9). Consensus sequences are generally stored in widely used databases such as Repbase (18) or along with seed alignments and HMMs in Dfam (19). Seed alignments and accurate sequence models are critical for reconstructing the evolutionary history of TEs and are used for a variety of biological studies, including the study of TE invasions and regulation (e.g., ref. 20). Years of manual curation have resulted in high-quality consensus libraries for a limited set of species, mostly model organisms (19, 21, 22).
The number of whole-genome assemblies for species throughout the tree of life continues to grow at a very fast rate, and efforts are underway to produce thousands more (23, 24). Long-read sequencing technologies are improving the quality of genome assemblies, especially in highly repetitive regions (e.g., ref. 25). These developments bring a pressing need to improve tools that automate the discovery and annotation of TEs. Although there are dozens of tools that tackle one aspect of de novo identification or one class of TE (26, 27), there are very few easy-to-use programs that can produce a comprehensive library of TE family seed alignments and consensus sequences from a genome assembly.
RepeatModeler was released in 2008 by Hubley and Smit and is one of the most widely used TE discovery tools (cited 1,462 times in publications as of 21 November 2019). RepeatModeler constructs seed alignments and consensus sequences for genome-wide repeat families de novo. However, the original version of RepeatModeler, like other existing TE discovery software, falls short of producing a complete, nonredundant library of full-length consensus sequences. The most problematic issue is the resolution of what should be a unique contiguous consensus sequence for a given TE family from many fragmented and partially redundant sequences in the output library. This issue, in turn, can hamper the classification of the TE families, inflates the number of actual TE families in the genome, and confounds genome annotation and downstream analyses. LTR retroelements are particularly recalcitrant to automated TE identification because of their size (up to 20 kilo base pairs [kbp]) and complexity in sequence and organization, which is driven, in part, by their ability to recombine within and between families (28). Yet these elements are widespread and often extremely abundant and diverse in eukaryotic genomes. For instance, the maize reference genome harbors >100,000 LTR elements falling into ∼20,000 distinct families accounting for about half of the genomic DNA (29).
To address these issues, we developed an improved version of RepeatModeler. Notably, we integrated an optional module dedicated to the identification of LTR elements in the genome through their structural characteristics (30, 31). By benchmarking on three diverse model species, we demonstrate that RepeatModeler2 is a substantial improvement over the previous version both in terms of detection sensitivity and consensus sequence quality. The open-source package is designed to run on a single, multiprocessor computer and is available as a source distribution or Docker/Singularity container for easier installation (https://github.com/Dfam-consortium/RepeatModeler).
Methods
RepeatModeler2 Overview.
RepeatModeler is a pipeline for automated de novo identification of TEs that employs two distinct discovery algorithms, RepeatScout (32) and RECON (33), followed by consensus building and classification steps. In addition, RepeatModeler2 now includes the LTRharvest (30) and LTR_retriever (34) tools. Our tool takes advantage of the unique strengths of each approach as well as providing a tractable solution to analyzing large datasets such as whole-genome assemblies. For instance, RepeatScout uses high-frequency sequence word counts to identify interspersed repeat seeds (short regions of putative homology) and then performs an iterative extension of a multiple alignment around the aligned seeds, similar to the seed and extend phase of the BLAST pairwise alignment algorithm. While RepeatScout’s implementation of this algorithm is fast, the program input is currently limited to ∼1 Gbp and the alignment scoring system (+1/−1, and nonaffine gap penalty) limits the divergence of discovered families. Despite these limitations, RepeatScout serves well as a fast method to discover the youngest and most abundant families given a small sequence sample from a genome. RECON, on the other hand, provides sophisticated and TE biology-aware clustering and relationship determination approaches to generate TE families from exhaustive intergenome alignments. RECON’s approach requires a computationally intensive but sensitive alignment (sophisticated scoring matrices, and affine gap penalties) and detects older TE families quite well.
In order to comprehensively identify TE families in a genome, we chose to employ a sampling and iterative (sample, mask, identify) search strategy (Fig. 1). We begin by supplying RepeatScout with a random 40-Mbp sample of the genome to quickly identify young and abundant families. In each successive round, we mask a new genomic sample using all previously discovered TE families to avoid rediscovery and allowing for larger successive sample sizes as the computational burden of self-comparison is reduced on the premasked sequence. The second and subsequent rounds all employ the RECON approach starting with a 3-Mbp sample (without replacement), tripling the sample size between rounds, and continuing until a sample size maximum or round limit is reached (default: 243 Mbp, or five rounds). For an average mammalian genome of 3 Gbp, this would sample ∼13% of the genome.
Fig. 1.
RepeatModeler2 flow diagram.
The new RepeatModeler2 pipeline now includes an additional structural discovery approach to assist in the discovery of LTR retrotransposons. Due to their unique structure and biology (two LTRs flanking a large 5- to 18-kb internal region), LTRs are often identified as fragments with disassociated LTR and internal regions or missing the internal segment completely using the RepeatScout/RECON methodologies. The LTR discovery is run on the complete unmasked genomic sequence and, as such, produces high-quality LTR families with some redundancy to the previously discovered families. Therefore, we follow up LTR discovery with a merging and redundancy removal process as described in LTR Module Description.
Due to constraints in the methods employed by TE discovery algorithms, their output often either is in the form of a complete or partial set of genome annotations (location ranges within the input sequence representing instances of a particular repeat) or is simply the precalculated consensus for each discovered TE family. In addition, the quality of these precalculated consensus sequences may vary considerably. A basic goal of RepeatModeler has been to produce a high-quality seed alignment and consensus sequence for each TE family; therefore we developed a seed alignment refinement method (see Refiner Description) which is employed on all families produced by the de novo tools.
Once all rounds of discovery/refinement/merging are complete, the final library is run through a simple classification step (see Classification for details), where each family is assigned (if possible) to a known TE class using the unified RepeatMasker/Dfam classification nomenclature.
Refiner Description.
Given a set of putatively related TE family instances, the RepeatModeler Refiner tool attempts to build a high-quality seed alignment and derive from it a consensus sequence for the family (SI Appendix, Fig. S1). Refiner bootstraps the generation of a seed alignment by selecting, as the initial consensus, the sequence which scores the best against all others. From this initial alignment, a new consensus is generated, and the process is repeated until the consensus stabilizes. Refiner employs an algorithm to identify low-scoring subregions in the seed alignment, often caused by common indels relative to the consensus, and resolves them by globally aligning the sequences within the subregion and updating the consensus (SI Appendix). The consensus is not simply based on alignment column majority rule; rather, each consensus position represents the highest-scoring base using a scoring matrix similar to those developed for RepeatMasker, which reflects observed neutral substitution patterns in mammals (SI Appendix, Fig. S2). For instance, the algorithm is aware of the rapid decay of CpG sites to CpA and TpG dinucleotides in most eukaryotes due to accelerated deamination of methylated cytosines (35, 36) and calls a CG pair given enough instances of aligned CA and TG dinucleotides (SI Appendix, Table S1). The final output of the analysis is a consensus sequence and a seed alignment in Stockholm format. The latter can be used for generating profile HMMs (37) and preserves the provenance of the family’s representative sequences.
LTR Module Description.
RepeatModeler2 uses the LTRharvest (30) package for structural LTR detection for both its overall sensitivity and speed (31, 38). LTRharvest is both a discovery and annotation algorithm that does not attempt to group LTR instances into families. In addition, any region in the genome demonstrating LTR-like structure (flanking repetitive sequence of the correct size, with the correct intervening sequence) is often incorrectly identified as an LTR instance. To solve this problem, Ou and Jiang (34) developed LTR_retriever, a package for filtering false positive results, resolving nested (mosaic) annotations, and identifying internal regions of LTRs. Some genomes have challenging nested structures that are not always resolved by LTR_retriever, so we implemented an optional parameter (-LTRMaxSeqLen) that sets the maximum allowable LTR internal length to avoid inclusion of missed mosaic internal elements in the seed alignment (SI Appendix).
We use LTR_retriever’s redundant library and perform our own clustering and consensus-building process. Since LTR elements frequently contain internal deletions, and this often results in “oversplitting” elements when clustering with CD-HIT (39), we implemented a clustering approach that scores alignment gaps as a reduced (single position) penalty. This step involves a multiple sequence alignment with MAFFT (40), followed by nearest-neighbor clustering into families with Ninja (41). Families are then refined and consensus sequences generated in a similar fashion to results from RepeatScout/RECON.
Combining Libraries and Reducing Redundancy.
Combining results from multiple tools is a difficult but essential step for the production of a comprehensive and nonredundant library of TE families. The RepeatScout and RECON analysis rounds reduce redundancy by masking out previously identified families with each iteration. With the addition of the LTR structural module as an independent analysis on the genome, we cannot avoid generating redundant LTR TE families. We tackled this problem by clustering the sequences between the modules with CD-HIT. Whenever RepeatModeler sequences cluster with an LTR pipeline sequence, we retain the LTR pipeline family as the representative. In addition, in RepeatModeler2, we extended this method to RepeatScout/RECON-produced families by labeling closely matching sequences as putative subfamilies with a reference to the accepted family representative. Users can then choose whether to remove these subfamilies, depending on the goals of their analyses.
Classification.
RepeatModeler contains a basic homology-based classification module (RepeatClassifier) which compares the TE families generated by the various de novo tools to both the RepeatMasker Repeat Protein Database (DB) and to the RepeatMasker libraries (e.g., Dfam and/or RepBase). The Repeat Protein DB is a set of TE-derived coding sequences that covers a wide range of TE classes and organisms. As is often the case with a search against all known TE consensus sequences, there will be a high number of false positive or partial matches. RepeatClassifier uses a combination of score and overlap filters to produce a reduced set of high-confidence results. If there is a concordance in classification among the filtered results, RepeatClassifier will label the family using the RepeatMasker/Dfam classification system and adjust the orientation (if necessary). Remaining families are labeled “Unknown” if a call cannot be made. Classification is the only step that requires a database, and can be completed with only open-source Dfam if Repbase is not available.
Benchmarking.
We benchmarked RepeatModeler2 on model species that have high-quality reference TE libraries: Drosophila melanogaster, Danio rerio, and Oryza sativa. We used Repbase for the D. melanogaster (release 20181026) and D. rerio (release 14.01) libraries, and used the manually improved library for O. sativa from ref. 38. Details required to reproduce the benchmark libraries are provided in SI Appendix, Table S2. We used RepeatMasker and parseRM (https://github.com/4ureliek/Parsing-RepeatMasker-Outputs/blob/master/parseRM.pl) to estimate the percentage of the genome masked by each subclass for the manually curated and RepeatModeler2 libraries.
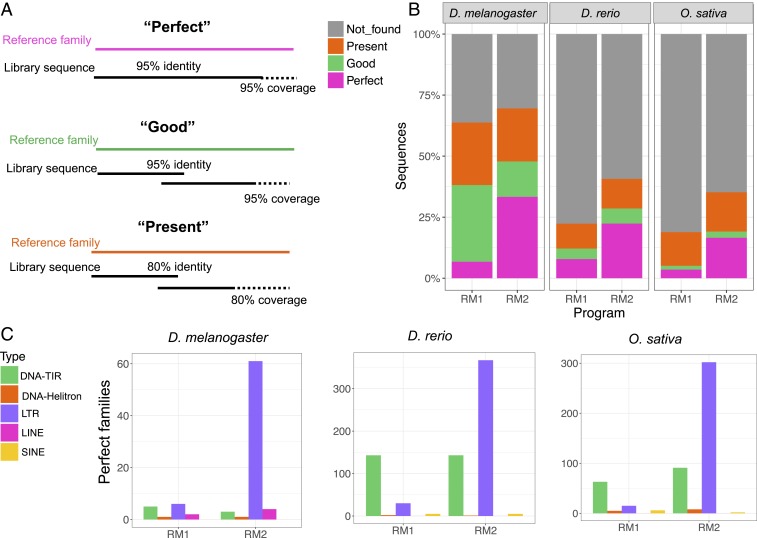
An important aspect of our pipeline is its ability to produce accurate consensus sequences corresponding to unique TE families. Thus, we also assessed the quality of our families by comparing their consensus sequences with the sequences of the manually curated reference libraries. We used RepeatMasker (4.0.8) to annotate the RM2 consensus sequences with the curated reference libraries and a custom bash script (https://github.com/jmf422/TE_annotation/blob/master/get_family_summary_paper.sh) to assess the sequence identity and coverage of matches between the libraries. Families were classified as being “perfect,” “good,” “present,” or “not found” based on the following definitions. “Perfect” families are those for which one sequence in our de novo library matches with >95% nucleotide similarity and >95% length coverage to a family consensus in the reference library. “Good” families are those in which multiple overlapping sequences in our output library match with >95% nucleotide similarity and >95% coverage to the curated consensus. A family is considered “present” if one or multiple library sequences align with >80% similarity and >80% coverage to the reference consensus sequence. Below these thresholds, we consider a family “not found” (although there may be fragments present in our output library).
Data Availability.
The RepeatModeler2 software is available at https://github.com/Dfam-consortium/RepeatModeler and http://www.repeatmasker.org/RepeatModeler/. Questions regarding the software should be directed to R.H. ([email protected]). Scripts used for evaluating the benchmarking results are available at https://github.com/jmf422/TE_annotation/. The TE libraries produced by RepeatModeler1 and RepeatModeler2 that were used for benchmarking are available at https://github.com/jmf422/TE_annotation/tree/master/benchmark_libraries.
Results and Discussion
We benchmarked RepeatModeler2 on three model species (fruit fly, zebrafish, rice) with diverse TE landscapes for which reference TE libraries have been extensively curated over decades of study (Fig. 2). As a first assessment of the ability of RepeatModeler2 to capture known TEs in each of these genomes, we used each output library to run RepeatMasker against the cognate genome assembly and measured the percent of the genome masked by each major TE subclass. We also counted the number of families in each library assigned to each subclass. RepeatModeler2 was able to recover the contrasted TE landscapes of these species (compare with curated libraries in Fig. 2).
Fig. 2.
Benchmarking of RepeatModeler2 on three model species. (Top) Genome composition (Upper) and number of families (Lower) of each TE subclass for the reference libraries. (Bottom) Genome composition (Upper) and number of families (Lower) of each TE subclass for the RepeatModeler2 library.
RepeatModeler2 produced similar TE composition profiles to the reference libraries. As previously documented (e.g., ref. 21), our results show that the genome of the fruit fly D. melanogaster is dominated by retrotransposons, especially LTR retroelements. This is reflected both by the amount of genomic DNA covered by these elements and by the number of unique families (Fig. 2). The zebrafish, D. rerio, is dominated by class II, DNA-TIR transposons, but also exhibits a very diverse assortment of LTR retroelements with many unique families (42). Our RepeatModeler2 library captures this general composition, but with a slight overrepresentation of DNA-TIR elements. This is likely caused by multiple consensus sequences representing fragments from the same family, which is sometimes produced by the RECON/RepeatScout module of RepeatModeler2. The genome of rice, O. sativa, is known to contain almost equal proportions and numbers of DNA-TIR and LTR elements (38), and this profile is recovered by our RepeatModeler2 library (Fig. 2). In summary, RepeatModeler2 produces libraries that recapitulate the major TE subclass composition of these three model species.
Next, we assess the ability of RepeatModeler2 to accurately capture the diversity and sequence of unique TE families. RepeatModeler2 produced 766, 3,851, and 2,648 library consensus sequences for D. melanogaster, D. rerio, and O. sativa, respectively—all comparable to the number of individual sequences in the reference libraries (Table 1). In addition, RepeatModeler2 labels families that cluster within 20% similarity as “putative subfamily”; thus we also provided the number of sequences without the inclusion of subfamilies (Table 1). A TE library with more sequences is not necessarily more useful, since it often indicates redundancy and fragmented sequences. Since we use a redundancy removal step, RepeatModeler2 did not produce drastically more family models than the previous version of the program.
Table 1.
Number of sequences produced from RepeatModeler and present in the manually curated libraries
| Species | RM2 families | RM2 families (no subfamilies) | Curated library | RM1 families |
| D. melanogaster | 734 | 509 | 207 | 699 |
| D. rerio | 3,851 | 3,147 | 2,322 | 2,503 |
| O. sativa | 2,648 | 2,284 | 2,431 | 1,652 |
The second column indicates the total number of families produced by RepeatModeler2, and the third column is the same except not including those annotated as subfamilies. The fourth column is the number of sequences in the curated library, and the fifth column is the number of sequences produced from RepeatModeler1.
The most significant improvement of RepeatModeler2 over the previous release of the program is in the quality (accuracy) of the family consensus sequences delivered in the output library (Fig. 3). We labeled the sequence matches between the RepeatModeler2 and reference libraries as “perfect,” “good,” or “present” based on the level of sequence similarity and coverage (Fig. 3A; see also Methods). If the family did not meet the minimum criteria for being counted as “present,” it was reported as “not found.” RepeatModeler2 produced 2.9 to 4.7 times more “perfect” families than the first version of RepeatModeler, and had more consensus sequences closely matching the reference libraries overall (Fig. 3B). Most of this improvement can be attributed to perfect LTR element families that are identified by RepeatModeler2 but were previously missed (Fig. 3C). The evaluation criteria we used for families were relatively strict, probably explaining the large number of families “not found.” Indeed, when masking each genome with the cognate library, we were able to recapitulate the genomic proportions of each subclass as generally obtained with the reference library (Fig. 2).
Fig. 3.
Evaluation family by family for RepeatModeler1 and RepeatModeler2. (A) Definitions of “Perfect,” “Good,” and “Present” families. “Perfect” families are those for which one sequence in our de novo library matches >95% in sequence identity and coverage to a family in the reference library. “Good” families are those in which multiple overlapping library sequences with alignments >95% similar to the reference consensus make up the >95% sequence coverage of the element. Finally, a family is considered “present” if one or multiple library sequences align with >80% similarity to the reference consensus sequence and cover >80% of the sequence. Otherwise, we consider a family “not found” (although there may be fragments present). (B) Summary of families found by the last release of RepeatModeler (RM1) and RepeatModeler2 (RM2). (C) Number of perfect families by subclass for each benchmark species.
Eukaryotic genomes contain complex structure and tandem repeats, which may result in false positives for TE discovery software. We assessed the false positive rate of RepeatModeler2 by running it on artificially generated genomes devoid of TEs simulated by GARLIC (43) for D. melanogaster and D. rerio. GARLIC generates background sequences with realistic complexity, isochore structure, and tandem repeat content similar to the modeled genome. RepeatModeler2 produced only one false positive family on the D. melanogaster artificial genome and five false positive families on the D. rerio artificial genome. No false positive families were produced from the LTR module. These results suggest that the rate of false positives generated by RepeatModeler2 is very low.
In addition to LTR retroelements, other types of TEs are sometimes considered challenging to identify ab initio with automated approaches. For example, short, noncoding DNA-TIR elements, often called MITEs (miniature inverted-repeat TEs), may be difficult to build because of their short length and sometimes palindromic nature (44, 45). However, the large number of well-characterized MITEs in the rice genomes (46) did not appear underrepresented in the RepeatModeler2 libraries compared to their long, coding peers (53% and 32% of families found for MITEs and coding DNA-TIRs, respectively). In comparison to the reference libraries, RepeatModeler2 also performed well with the identification of Helitrons, which also pose particular challenges for automated discovery (reviewed in ref. 14) (Fig. 2). There are still limitations on detecting some types of elements that do not have dedicated structural detection tools, such as Polinton/Maverick elements. However, because the RECON/RepeatScout module is agnostic to different TE structures and only relies on the nature of interspersed repeats, it can detect TEs with more atypical structures. Thus, RepeatModeler2 appears capable of recovering a wide diversity of TEs which have been traditionally considered recalcitrant to ab initio identification.
Several programs exist that identify signatures of repetitive sequences in genomes (e.g., ref. 47); however, few attempt to generate a library of TE family models suitable for high-quality genome annotation. The former do not typically attempt to identify or model the individual repeating units or distinguish between low-complexity, tandem, or transposable (interspersed) repeats.
These methods are useful for screening out repetitive DNA, but do not directly assist with the development of TE libraries or facilitate the detailed annotation of a genome. We compared RepeatModeler2 to other TE annotation programs which produce TE libraries, REPET (48) and EDTA (38). RepeatModeler2 found more “perfect” sequences than both REPET and EDTA on D. melanogaster (SI Appendix, Fig. S3). However, REPET found more families overall. EDTA (which was developed using rice as the benchmark) found more families (“perfect,” “good,” and “present”) in rice than RepeatModeler2 (SI Appendix, Fig. S3). Both EDTA and REPET produced about 10-fold more consensus sequences than RepeatModeler2, making the ratio of number of curated families to number of library consensus sequences much lower (SI Appendix, Fig. S4). A large number of library sequences indicates fragmentation, false positives, and redundancy, which can be difficult to manage in downstream analysis. Depending on the goals of analysis, these other tools may be complementary to use alongside RepeatModeler2.
We anticipate that additional improvements will further enhance the current version of the pipeline. Because of the modular architecture of RepeatModeler2, it should be relatively straightforward to add other modules tailored to the discovery of specific subclasses of elements such as those dedicated to the identification of MITEs (44), or Helitrons (49, 50). It is also important to emphasize that the classifier currently used by RepeatModeler2 remains rudimentary, as it strictly relies on detection of sequence homology to known TEs and protein domains. This limitation hampers the ability to classify noncoding elements or elements with sequences highly diverged from those annotated in the databases. Integrating additional features used for TE classification, such as conserved TIR sequence motifs or target site duplications, as implemented in other TE classifiers (51) will further improve the ability of RepeatModeler2 to deliver high-quality libraries.
The genome annotation community is in pressing need of a TE discovery program that is easy to use, has been thoroughly benchmarked, and can be applied to almost any eukaryotic species (52). We believe that RepeatModeler2 will meet this demand. RepeatModeler2 is easy to install and run, as we provide a container version to avoid installing, independently, all dependencies. We anticipate that the application of RepeatModeler2 to existing and future genome assemblies will result in more-consistent genome annotations and improved TE family models, which will enhance a wide array of genomic analyses including but not limited to TE biology.
Supplementary Material
Acknowledgments
We thank Arnie Kas, Warren Gish, Alkes Price, Pavel Pevzner, Shujun Ou, and Ning Jiang for assistance with dependencies used by RepeatModeler. We thank Andy Siegel for statistics consultations in the development of RepeatModeler. This work was supported by NIH Grants U01-HG009391 and R35-GM122550 (to C.F.) and National Human Genome Research Institute Grants U24 HG010136 and R01 HG002939 (to A.F.S.). J.M.F. was supported by a Natural Sciences and Engineering Research Council of Canada Postgraduate Scholarship–Doctoral and National Institute of General Medical Sciences R01 Grant GM119125 to A.G.C.
Footnotes
Competing interest statement: C.F. and M.C.G.H. are coauthors on a 2018 review article: https://genomebiology.biomedcentral.com/articles/10.1186/s13059-018-1577-z.
Data deposition: The RepeatModeler2 software is available at https://github.com/Dfam-consortium/RepeatModeler, and http://www.repeatmasker.org/RepeatModeler/. Scripts used for evaluating the benchmarking results are available here: https://github.com/jmf422/TE_annotation/. The TE libraries produced by RepeatModeler1 and RepeatModeler2 that were used for benchmarking are available here: https://github.com/jmf422/TE_annotation/master/benchmark_libraries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1921046117/-/DCSupplemental.
References
- 1.Smit A. F., Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr. Opin. Genet. Dev. 9, 657–663 (1999). [DOI] [PubMed] [Google Scholar]
- 2.Lander E. S., et al. ; International Human Genome Sequencing Consortium , Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Huang C. R. L., Burns K. H., Boeke J. D., Active transposition in genomes. Annu. Rev. Genet. 46, 651–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourque G., et al. , Ten things you should know about transposable elements. Genome Biol. 19, 199 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jurka J., Kapitonov V. V., Kohany O., Jurka M. V., Repetitive sequences in complex genomes: Structure and evolution. Annu. Rev. Genomics Hum. Genet. 8, 241–259 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Appels R., et al. ; International Wheat Genome Sequencing Consortium (IWGSC) , Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361, eaar7191 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Hua-Van A., Le Rouzic A., Maisonhaute C., Capy P., Abundance, distribution and dynamics of retrotransposable elements and transposons: Similarities and differences. Cytogenet. Genome Res. 110, 426–440 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Smit A., RepeatMasker genomic datasets. http://www.repeatmasker.org/genomicDatasets/RMGenomicDatasets.html. Accessed 31 October 2019.
- 9.Yandell M., Ence D., A beginner’s guide to eukaryotic genome annotation. Nat. Rev. Genet. 13, 329–342 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Wicker T., et al. , A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 8, 973–982 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Finnegan D. J., Eukaryotic transposable elements and genome evolution. Trends Genet. 5, 103–107 (1989). [DOI] [PubMed] [Google Scholar]
- 12.Piégu B., Bire S., Arensburger P., Bigot Y., A survey of transposable element classification systems—A call for a fundamental update to meet the challenge of their diversity and complexity. Mol. Phylogenet. Evol. 86, 90–109 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Feschotte C., Pritham E. J., DNA transposons and the evolution of eukaryotic genomes. Annu. Rev. Genet. 41, 331–368 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thomas J., Pritham E. J., Helitrons, the eukaryotic rolling-circle transposable elements. Microbiol. Spectr. 3, (2015). [DOI] [PubMed] [Google Scholar]
- 15.Grabundzija I., Hickman A. B., Dyda F., Helraiser intermediates provide insight into the mechanism of eukaryotic replicative transposition. Nat. Commun. 9, 1278 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kapitonov V. V., Jurka J., Rolling-circle transposons in eukaryotes. Proc. Natl. Acad. Sci. U.S.A. 98, 8714–8719 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arkhipova I. R., Using bioinformatic and phylogenetic approaches to classify transposable elements and understand their complex evolutionary histories. Mob. DNA 8, 19 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bao W., Kojima K. K., Kohany O., Repbase update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 6, 11 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hubley R., et al. , The Dfam database of repetitive DNA families. Nucleic Acids Res. 44, D81–D89 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kofler R., Hill T., Nolte V., Betancourt A. J., Schlötterer C., The recent invasion of natural Drosophila simulans populations by the P-element. Proc. Natl. Acad. Sci. U.S.A. 112, 6659–6663 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lerat E., Rizzon C., Biémont C., Sequence divergence within transposable element families in the Drosophila melanogaster genome. Genome Res. 13, 1889–1896 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stitzer M. C., Anderson S. N., Springer N. M., Ross-Ibarra J., The genomic ecosystem of transposable elements in maize. bioRxiv:10.1101/559922 (28 February 2019). [DOI] [PMC free article] [PubMed]
- 23.Lewin H. A., et al. , Earth BioGenome Project: Sequencing life for the future of life. Proc. Natl. Acad. Sci. U.S.A. 115, 4325–4333 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koepfli K.-P., Paten B., O’Brien S. J.; Genome 10K Community of Scientists , The Genome 10K Project: A way forward. Annu. Rev. Anim. Biosci. 3, 57–111 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang C.-H., Larracuente A. M., Heterochromatin-enriched assemblies reveal the sequence and organization of the Drosophila melanogaster Y chromosome. Genetics 211, 333–348 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saha S., Bridges S., Magbanua Z. V., Peterson D. G., Empirical comparison of ab initio repeat finding programs. Nucleic Acids Res. 36, 2284–2294 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saha S., Bridges S., Magbanua Z. V., Peterson D. G., Computational approaches and tools used in identification of dispersed repetitive DNA sequences. Trop. Plant Biol. 1, 85–96 (2008). [Google Scholar]
- 28.Vargiu L., et al. , Classification and characterization of human endogenous retroviruses; mosaic forms are common. Retrovirology 13, 7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiao Y., et al. , Improved maize reference genome with single-molecule technologies. Nature 546, 524–527 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ellinghaus D., Kurtz S., Willhoeft U., LTRharvest, an efficient and flexible software for de novo detection of LTR retrotransposons. BMC Bioinformatics 9, 18 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ou S., Chen J., Jiang N., Assessing genome assembly quality using the LTR Assembly Index (LAI). Nucleic Acids Res. 46, e126 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Price A. L., Jones N. C., Pevzner P. A., De novo identification of repeat families in large genomes. Bioinformatics 21 (suppl. 1), i351–i358 (2005). [DOI] [PubMed] [Google Scholar]
- 33.Bao Z., Eddy S. R., Automated de novo identification of repeat sequence families in sequenced genomes. Genome Res. 12, 1269–1276 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ou S., Jiang N., LTR_retriever: A highly accurate and sensitive program for identification of long terminal repeat retrotransposons. Plant Physiol. 176, 1410–1422 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sved J., Bird A., The expected equilibrium of the CpG dinucleotide in vertebrate genomes under a mutation model. Proc. Natl. Acad. Sci. U.S.A. 87, 4692–4696 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Colot V., Rossignol J. L., Eukaryotic DNA methylation as an evolutionary device. BioEssays 21, 402–411 (1999). [DOI] [PubMed] [Google Scholar]
- 37.Wheeler T. J., Eddy S. R., nhmmer: DNA homology search with profile HMMs. Bioinformatics 29, 2487–2489 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ou S., et al. , Benchmarking transposable element annotation methods for creation of a streamlined, comprehensive pipeline. Genome Biol. 20, 275 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li W., Godzik A., Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Katoh K., Misawa K., Kuma K., Miyata T., MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wheeler T. J., “Large-scale neighbor-joining with NINJA” in Algorithms in Bioinformatics (Lecture Notes in Computer Science, Springer, 2009), vol. 5724, pp. 375–389. [Google Scholar]
- 42.Howe K., et al. , The zebrafish reference genome sequence and its relationship to the human genome. Nature 496, 498–503 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Caballero J., Smit A. F. A., Hood L., Glusman G., Realistic artificial DNA sequences as negative controls for computational genomics. Nucleic Acids Res. 42, e99 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Han Y., Wessler S. R., MITE-hunter: A program for discovering miniature inverted-repeat transposable elements from genomic sequences. Nucleic Acids Res. 38, e199 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feschotte C., Wessler S. R., Zhang X., “Miniature inverted-repeat transposable elements and their relationship to established DNA transposons” in Mobile DNA II (ASM Press, 2002), pp. 1147–1158. [Google Scholar]
- 46.Jiang N., Feschotte C., Zhang X., Wessler S. R., Using rice to understand the origin and amplification of miniature inverted repeat transposable elements (MITEs). Curr. Opin. Plant Biol. 7, 115–119 (2004). [DOI] [PubMed] [Google Scholar]
- 47.Girgis H. Z., Red: An intelligent, rapid, accurate tool for detecting repeats de-novo on the genomic scale. BMC Bioinformatics 16, 227 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Flutre T., Duprat E., Feuillet C., Quesneville H., Considering transposable element diversification in de novo annotation approaches. PLoS One 6, e16526 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang L., Bennetzen J. L., Structure-based discovery and description of plant and animal Helitrons. Proc. Natl. Acad. Sci. U.S.A. 106, 12832–12837 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiong W., He L., Lai J., Dooner H. K., Du C., HelitronScanner uncovers a large overlooked cache of Helitron transposons in many plant genomes. Proc. Natl. Acad. Sci. U.S.A. 111, 10263–10268 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Feschotte C., Keswani U., Ranganathan N., Guibotsy M. L., Levine D., Exploring repetitive DNA landscapes using REPCLASS, a tool that automates the classification of transposable elements in eukaryotic genomes. Genome Biol. Evol. 1, 205–220 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hoen D. R., et al. , A call for benchmarking transposable element annotation methods. Mob. DNA 6, 13 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RepeatModeler2 software is available at https://github.com/Dfam-consortium/RepeatModeler and http://www.repeatmasker.org/RepeatModeler/. Questions regarding the software should be directed to R.H. ([email protected]). Scripts used for evaluating the benchmarking results are available at https://github.com/jmf422/TE_annotation/. The TE libraries produced by RepeatModeler1 and RepeatModeler2 that were used for benchmarking are available at https://github.com/jmf422/TE_annotation/tree/master/benchmark_libraries.