Spontaneous intracerebral hemorrhage (ICH) accounts for 15–20% of “acute stroke” with a significant morbidity and mortality 1. Following ICH, pathobiological processes (blood–brain barrier (BBB) damage, cerebral edema, and inflammation) are activated 1, 2. The molecular changes associated with these processes could be a target for therapeutic interventions or potential biomarkers.
Gene expression profiling of human tissues and experimental models of ICH have identified potential candidate genes 3. MicroRNAs (miRNAs) are important gene regulators and primarily identified in tissues encapsulated in microvesicles 2, 4. Following tissue injury, miRNAs are released into the extracellular space 4. Comparison of miRNAs from cerebrospinal fluid (CSF) and blood from the same patient is unique as CSF likely reflects the local events of injured brain tissue compared to miRNAs in the bloodstream.
This study was approved by the Ochsner Medical Center Institution Review Board. Patients with ICH confirmed on computed tomography (CT) scan who required a ventricular drain were consented for CSF and blood sample collection. We enrolled five male subjects with a median age of 54 years (range, 37–78 years). The basic clinical information is provided (Table S1). Blood samples (10 mL) were collected in sodium citrate tubes, and CSF samples (3–5 mL) were collected within 24 h of presentation. The study was designed such that each patient serves as an intrinsic control in comparing CSF and blood miRNA expression patterns.
Total RNA containing miRNAs was isolated from plasma and CSF (200 μL) for profiling of 782 known miRNAs (Exiqon). The real‐time PCR was run for 40 cycles in a CFX‐384 thermal cycler (Bio‐Rad, Hercules, CA, USA). Cycle threshold (Ct) data were obtained using the regression method. Data were normalized using the global mean and log2‐transformed using the GenEx software (Exiqon, Vedbaek, Denmark). Using a Ct value cutoff of 39, 423 miRNAs were reliably expressed in CSF and plasma. Of these, 128 miRNAs were detected in CSF, 14 miRNAs in plasma, and 281 miRNAs in both CSF and plasma. Differential miRNA expression analysis found 242 significantly expressed miRNAs (P < 0.05). Of these, 139 miRNAs were found at 2‐fold higher in CSF and 94 miRNAs at 2‐fold lower in CSF compared to plasma. When the P‐values were adjusted for multiple testing correction using the Benjamin–Hochberg method, 211 miRNAs were statistically significant (P < 0.05; Table S2).
The heat map of the top 25 significantly expressed miRNAs is shown with the corresponding fold change and significance value (Figure 1A, B). The highly expressed miR‐204‐5p (2075‐fold), miR‐125b‐5p (108‐fold), miR‐9‐5p (299‐fold), miR‐338‐3p (146‐fold), miR‐187‐3p (21‐fold), and miR‐9‐3p (42‐fold) in CSF are associated with the regulation of matrix metalloproteinase‐9 (MMP‐9), IL‐1β, IL‐6, occludin, and SELE, to name a few. In the case of miR‐204‐5p, miR‐9‐3p, and miR‐338‐3p, which target MMP‐9, their expressions in plasma were decreased, suggesting an increased expression and activation of pro‐MMP‐9 in plasma. However, the expression in CSF is decreased. In animal and human studies, elevated MMP‐9 in plasma during the acute phase of ICH was associated with increased BBB damage, hematoma growth, and perihematomal edema 5.
Figure 1.
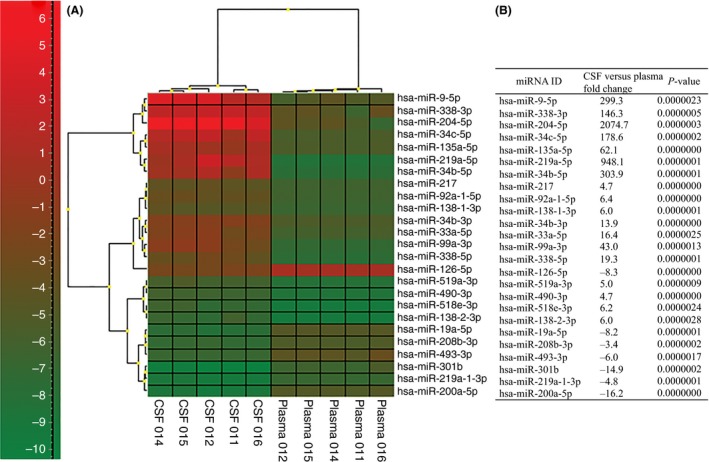
(A) Heatmap hierarchal cluster of the top 25 significantly expressed miRNAs; and (B) the top 25 significantly expressed miRNAs and the fold change in CSF relative to plasma. P‐values reported are with Benjamin–Hochberg multiple test correction.
MiR‐126‐5p expression was downregulated (−8.3‐fold) in CSF compared to plasma. miRNA‐126 regulates vascular cell adhesion molecule 1 (VCAM‐1) expression in endothelial cells. Decreased expression of miRNA‐126 may lead to VCAM‐1 expression and promotes immune cell localization at the site of injury. Zhu et al. 6 found miRNA levels in serum were mostly downregulated compared to healthy controls and that the serum level of miR‐126 correlated with perihematomal edema. On the contrary, Guo et al., 4 found upregulation of miRNAs associated with inflammation in plasma.
In healthy subjects, the numbers of miRNA in plasma are generally higher and unique compared with CSF 7. We found large increases in miRNA, both in abundance and fold change, in CSF than plasma. miRNA profile in CSF is physiologically contiguous with profiles in brain extracellular fluid 8. Hence, the differences in the miRNA content between CSF and plasma may reflect an altered systemic inflammatory response leading to neuroinflammation and BBB damage.
Pathway enrichment analysis of significant miRNAs using Diana‐miRPath v2.0 9 and the Kyoto Encyclopedia of Genes and Genomes (KEGG) identified pathways that may be activated in acute ICH. For the 128 miRNAs detected only in CSF, the top over‐represented KEGG pathways implicated in BBB disruption include PI3K‐Akt signaling, MAPK signaling, regulation of actin cytoskeleton, focal adhesion, endocytosis, TGF‐β signaling, gap, and adherens junctions. Predicted and validated target genes regulated by miRNAs were determined using the DIANA–TarBase v6.0 (Athena, Athens, Greece). Pathway enrichment analysis of the 139 miRNAs in CSF and 94 miRNAs in plasma found a similar pathway profile as those identified for CSF‐specific miRNAs (Table 1A). The miRNA‐regulated genes associated with these pathways were identified using the TarBase database of experimentally validated targets. Focal adhesion and neuroactive ligand receptor signaling were represented for upregulated miRNAs in CSF, whereas axon guidance and cytokine receptor signaling were represented by downregulated miRNAs. The validated targets for some of these miRNAs are associated with neuroinflammation (Table 1B). Previous studies have implicated the MAP kinase, Akt, and NFκβ signaling pathways and the involvement of these cytokines in neuroinflammation 5, 10. Although the miRNA profiles in CSF and plasma are distinct, the upregulated miRNAs in CSF and plasma regulate the same group of genes and shared many common signaling pathways associated with neuroinflammation and BBB damage. Therefore, systemic and intracerebral pathological conditions are reflective of one another, and that systemic monitoring may help guide intervention for brain injury after ICH. Hence, our study suggests a possible role for miRNA in the genetic regulation of these pathways and cytokines. Given the small sample size, we are not able to account for potential variations in miRNA profiles due to race, gender, or differences in healthy individuals.
Table 1.
Significantly expressed miRNAs found in CSF and plasma and their associated KEGG pathways. (A) The numbers of miRNAs that were found significantly expressed versus the total number of miRNAs associated with a KEGG pathway; (B) mRNA targets of the significantly expressed miRNAs and fold change relative to plasma
| A. Pathways and significantly expressed miRNAs | ||
|---|---|---|
| Pathwaya | miRNA in plasmab | miRNA in CSFb |
| Adherens junctions | 16/142 | 17/142 |
| MAPK signaling | 33/116 | 31/116 |
| WNT signaling | 22/104 | 26/104 |
| PI3K‐Akt signaling | 35/197 | 39/197 |
| Focal adhesion | 82/1593 | 105/1593 |
| Actin cytoskeleton | 22/85 | 23/85 |
| Apoptosis | 23/142 | 24/142 |
| Cytokine receptor signaling | 27/98 | 19/98 |
| Neuroactive ligand receptor signaling | 4/35 | 10/35 |
| Axon guidance | 20/664 | 13/664 |
| Ubiquitin‐mediated proteolysis | 14/60 | 14/60 |
| Dopaminergic signaling | 16/65 | 17/65 |
| B. mRNA target: miRNA (Fold change) | ||
| MMP‐9: miR‐204‐5p (2075); miR‐338‐3p (146);miR‐9‐5p (299); mir‐211‐5p (15); miR‐29b‐3p (6); let‐7e‐5p(6) | ||
| TNF‐α: miR‐187‐3p (21);miR‐143‐3p (2); miR‐130a‐3p (2) | ||
| IL‐1β: miR‐204‐5p (2075) | ||
| IL‐6: miR‐125b‐5p (108) | ||
| Occludin: miR‐9‐3p (42); miR‐488‐3p (12) | ||
| SELE: miR‐204‐5p (2075); miR‐211‐5p (15) | ||
| CCR5: miR‐125b‐5p (108) | ||
| ICAM‐1: miR‐187‐3p (21); miR‐155‐5p (‐13); miR‐31‐5p (3); miR‐221‐3p (‐3); miR‐222‐3p (‐3) | ||
| CXCL8: miR‐302a‐3p (21) | ||
| CXCL5: miR‐488‐3p (13); miR‐577 (16) | ||
| CXCL10: let‐7a‐2‐3p (12) | ||
aPathways that were found significant based on a threshold cutoff (P < 0.05) with FDR correction using Diana‐TarBase v6.0 and miRPath default setting. bThe number of input miRNA to the number of miRNA represented in the respective KEGG pathway Akt, Ak strain transforming; KEGG, Kyoto Encyclopedia of Gene and Genome; MAPK, mitogen‐activated protein kinase; miRNA, microRNA; PI3K, phosphoinositide‐3‐kinase; Wnt, wingless‐type MMTV integration site family, CXCL5, chemokine (C‐X‐C motif) ligand 5; CXCL8, chemokine (C‐X‐C motif) ligand 8 (Interleukin 8); CXCL10, chemokine (C‐X‐C motif) ligand 10; ICAM‐1, intercellular adhesion molecule‐1; IL‐1β, interleukin‐beta‐1; IL‐6, interleukin‐beta‐6; MMP‐9, metalloproteinase‐9; SELE, selectin E.
In conclusion, the present study identified a significantly greater number of miRNAs in the CSF than plasma of ICH patients. The large fold decrease in the plasma miRNAs that regulate inflammation‐associated genes suggests that the systemic regulation of these genes may be affected. These miRNAs can provide insights on the underlying neuroinflammation in ICH.
Conflict of interests
The authors declare that they have no competing interests. No portion of this study has been published or is under consideration for publication elsewhere. The authors have no personal, financial, or institutional interests in any of the drugs and materials described in this manuscript.
Supporting information
Table S1. Patient basic clinical information.
Table S2. Significantly expressed miRNA in CSF vs plasma.
Acknowledgments
We thank Dr. Walter Lukiw for the scientific review of this manuscript and thank Lauren Allulli and Michelle Beavers for patient consent, sample preparation, and manuscript review. Research reported in this publication/presentation is supported by the Ochsner Translational Medicine Research Initiative sponsored by the Ochsner Health System (II), and the Ochsner Clinic Foundation (II and WS).
References
- 1. Keep RF, Hua Y, Xi G. Intracerebral haemorrhage: mechanisms of injury and therapeutic targets. Lancet Neurol 2012;11:720–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zheng HW, Wang YL, Lin JX, Li N, Zhao XQ, Liu GF. Circulating MicroRNAs as potential risk biomarkers for hematoma enlargement after intracerebral hemorrhage. CNS Neurosci Ther 2012;18:1003–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yang T, Gu J, Kong B, et al. Gene expression profiles of patients with cerebral hematoma following spontaneous intracerebral hemorrhage. Mol Med Rep 2014;10:1671–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Guo D, Liu J, Wang W, et al. Alteration in abundance and compartmentalization of inflammation‐related miRNAs in plasma after intracerebral hemorrhage. Stroke 2013;44:1739–1742. [DOI] [PubMed] [Google Scholar]
- 5. Florczak‐Rzepka M, Grond‐Ginsbach C, Montaner J, Steiner T. Matrix metalloproteinases in human spontaneous intracerebral hemorrhage: An update. Cerebrovasc Dis 2012;34:249–262. [DOI] [PubMed] [Google Scholar]
- 6. Zhu Y, Wang JL, He ZY, Jin F, Tang L. Association of altered serum microRNAs with perihematomal edema after acute intracerebral hemorrhage. PLoS One 2015;10:e0133783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Webber JA, Baxter DH. The microrna spectrum in 12 bodily fluids. Clin Chem 2010;56:11 (1733‐1741). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Alexandrov PN, Dua P, Hill JM, Bhattacharjee S, Zhao Y, Lukiw WJ. microRNA (miRNA) speciation in Alzheimer's disease (AD) cerebrospinal fluid (CSF) and extracellular fluid (ECF). Int J Biochem Mol Biol 2012;3:365–373. [PMC free article] [PubMed] [Google Scholar]
- 9. Vlachos IS, Kostoulas N, Vergoulis T, et al. DIANA miRPath v.2.0: Investigating the combinatorial effect of microRNAs in pathways. Nucleic Acids Res 2012;40(Web Server issue):W498–W504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ohnishi M, Monda A, Takemoto R, et al. Sesamin suppresses activation of microglia and p44/42 MAPK pathway, which confers neuroprotection in rat intracerebral hemorrhage. Neuroscience 2013;232:45–52. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Patient basic clinical information.
Table S2. Significantly expressed miRNA in CSF vs plasma.


