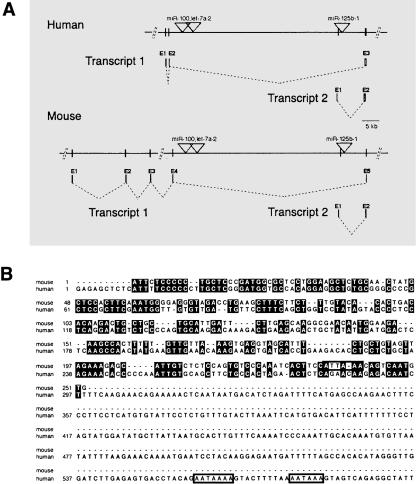
Figure 1.
Genomic organization of the mammalian miR-100, let-7a-2, miR-125b-1 locus and sequence alignment of human and mouse miR-125b-1 host transcripts. (A) Identified transcripts are shown with exons represented as vertical lines and labeled above in numerical order. Dotted lines denote splicing of transcripts. Positions of miRNA precursors are denoted with a triangle. Exons are not to scale. Human/Mouse Transcript 1 corresponds to ENSESTT00000036713 and ENSMUSESTT00000030830, respectively. Transcript 2 corresponds to human ENSESTT00000036715 and mouse 2610203C20Rik transcripts. (B) A DNA sequence alignment for Transcript 2 is shown. Identical bases are denoted in black. Consensus polyadenylation sites are boxed in. Note that nucleotides 597-955 of the human Transcript 2 are not shown.