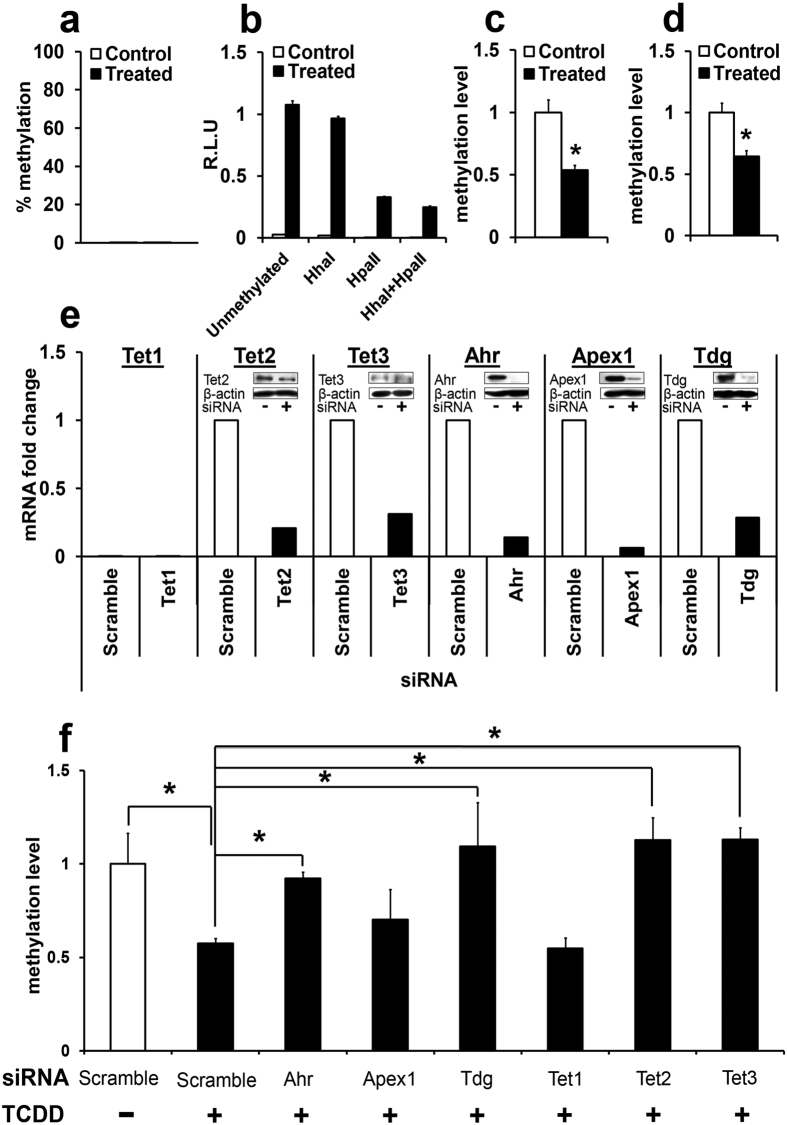
Figure 5. Ahr, Tdg, Tet2, and Tet3 are required for Ahr-directed DNA demethylation.
(a) Hepa1c1c7 endogenous Cyp1a1 promoter methylation levels at the target CpGs as analyzed by MSRE-qPCR. (b) The in vitro methylated Cyp1a1 reporter plasmid response to TCDD as analyzed by luciferase expression. 100 ng of the methylated plasmid was transfected into Hepa1c1c7 cells. After treatment with 10 nM TCDD for 24 hrs, luciferase activity was measured by the Dual-Luciferase® Reporter Assay System. (c,d) methylation levels of the −500 and −420 CpG sites respectively, in the in vitro methylated plasmids after TCDD exposure. Data are expressed as mean ± SE. (n = 3). *P < 0.05, Student’s t-test. (e) siRNA knockdown efficiency 48 hrs after transfection; mRNA levels were measured by RT-qPCR and protein levels by western blot. (n = 2). (f) Methylation level of the methylated plasmid after siRNA target knockdown and TCDD treatment. Data are expressed as mean ± SE. (n = 3). *P < 0.05, one-way ANOVA followed by Fisher's PLSD post hoc test.