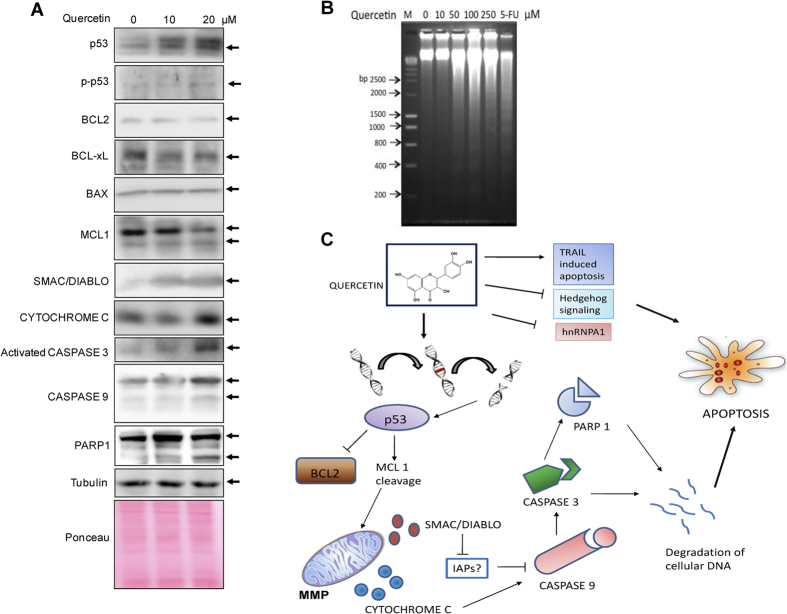
Figure 8. Effect of quercetin on expression of apoptotic proteins and mechanism of action.
(A) Expression level of apoptotic proteins following treatment with quercetin. Cell lysate was prepared after treating Nalm6 cells with quercetin (0, 10, 20 μM, for 24 h). Protein lysate was resolved on SDS-PAGE and immunoblotting was performed using appropriate primary and secondary antibodies. Expression pattern was studied for p53, p-p53, BCL2, BCL-xL, BAX, MCL 1, CYTOCHROME C, SMAC/DIABLO, activated CASPASE 3, CASPASE 9, PARP1 and Tubulin. Ponceau stained PVDF membrane after protein transfer acted as a control for equal protein loading. (B) Detection of DNA fragmentation in Nalm6 cells following treatment with quercetin. Chromosomal DNA was extracted after treatment with quercetin (10, 50, 100, 250 μM). The purified DNA was resolved on 2% agarose gel at 50 V for 5 h. M is hyper ladder I (Bioline). 5-fluorouracil (5-FU; 100 μM) was used as positive control. (C) Proposed model for quercetin induced apoptosis leading to cytotoxicity. Treatment of cancer cells with quercetin leads to apoptosis through activation of mitochondrial intrinsic pathway. Since quercetin is a DNA intercalator, it might induce DNA damage, which leads to the upregulation of p53. This results in the down regulation of antiapoptotic protein, BCL2 and cleavage of MCL1. CYTOCHROME C and SMAC/DIABLO are released due to loss in mitochondrial membrane potential. SMAC/DIABLO inhibits IAPs (inhibitors of apoptosis proteins), while CYTOCHROME C release leads to cleavage of CASPASE 9, which in turn cleaves CASPASE 3. CASPASE 3 leads to fragmentation and degradation of cellular DNA and activation of PARP1 resulting in apoptosis.