ABSTRACT
Coronaviruses are RNA viruses with a large zoonotic reservoir and propensity for host switching, representing a real threat for public health, as evidenced by severe acute respiratory syndrome (SARS) and the emerging Middle East respiratory syndrome (MERS). Cellular factors required for their replication are poorly understood. Using genome-wide small interfering RNA (siRNA) screening, we identified 83 novel genes supporting infectious bronchitis virus (IBV) replication in human cells. Thirty of these hits can be placed in a network of interactions with viral proteins and are involved in RNA splicing, membrane trafficking, and ubiquitin conjugation. In addition, our screen reveals an unexpected role for valosin-containing protein (VCP/p97) in early steps of infection. Loss of VCP inhibits a previously uncharacterized degradation of the nucleocapsid N protein. This inhibition derives from virus accumulation in early endosomes, suggesting a role for VCP in the maturation of virus-loaded endosomes. The several host factors identified in this study may provide avenues for targeted therapeutics.
IMPORTANCE Coronaviruses are RNA viruses representing a real threat for public health, as evidenced by SARS and the emerging MERS. However, cellular factors required for their replication are poorly understood. Using genome-wide siRNA screening, we identified novel genes supporting infectious bronchitis virus (IBV) replication in human cells. The several host factors identified in this study may provide directions for future research on targeted therapeutics.
INTRODUCTION
Coronaviruses (CoV) are a family of enveloped, positive-strand RNA viruses that can cause respiratory and enteric disease both in humans and in a wide range of animals. To date, six different virus species are known to affect humans (1). The severe acute respiratory syndrome (SARS) CoV is the etiological agent responsible for an outbreak in 2002 to 2003, which caused approximately 10% mortality in the 8,000 people infected worldwide (2). A recently emerged CoV is responsible for the recent outbreak of Middle East respiratory syndrome (MERS).
The other human coronaviruses (HCoV)—HCoV-229E, HCoV-OC43, NL63, and HKU1—are collectively responsible for about 10 to 30% of common colds. Generally harmless and self-limiting, these HCoV are also implicated in severe clinical outcomes, particularly in immunocompromised individuals, infants, and the elderly (3). Other coronaviruses cause considerable economic concern to the livestock industry as they readily infect farmed animals such as cows (4), pigs (5), and chickens (6).
In addition to the diverse range of species that they infect, coronaviruses have a propensity for host switching. For instance, HCoV-OC43 bears a strong resemblance to a bovine coronavirus, from which it probably originated (7). SARS-CoV is postulated to have originated from bats and then transferred to palm civets and finally humans (8). The MERS coronavirus probably also has its origin in bats and is responsible for severe respiratory and renal failure in humans (9). Although human-to-human transmission is low at present (10), this new beta-coronavirus has raised global health concerns because its mortality rate is more than 30% (1, 11). This “interspecies jumping” continuously threatens to initiate a novel epidemic and presents a challenge for vaccine-based containment.
It is thus critical to have a better understanding of the infectious cycle of CoV. This multistep process includes attachment of the spike protein (S) to cell surface receptors, endocytosis, and then fusion of the viral and endocytic membranes (12). The viral capsid then undergoes an uncoating process to deliver the viral genome into the cytosol.
Host ribosomes then translate the viral genome, yielding nonstructural proteins that modulate virus pathogenesis (13) and form with host membranes the viral transcription/replication complex (RTC). The RTC is responsible for transcription of full-length genomic RNA as well as subgenomic RNA species via a nidovirus-specific discontinuous transcription mechanism. Translation of the subgenomic RNAs produces viral structural proteins, nucleocapsid protein (N), membrane protein (M), envelope protein (E), and spike protein (S). Finally, structural proteins are packaged with genomic RNA into new virions secreted via the secretory pathway.
For all these steps, host proteins and cellular pathways are required. Drug inhibition and small interfering RNA (siRNA) studies have highlighted the early secretory pathway (14, 15) and DNA damage response (16). Direct interactions between host and viral proteins aid in virus assembly (16, 17). Recently, a genome-wide yeast two-hybrid screen revealed the involvement of cyclophilins and the calcineurin/NFAT (nuclear factor of activated T cells) pathway in SARS-CoV replication (18). Conversely, coronavirus proteins can inhibit cellular processes that may impede its replication, such as the interferon response (19).
Targeting host factors may provide broad-specificity viral inhibitors, but an intimate knowledge of the required host genes is a prerequisite. In this paper, we describe the use of a genome-wide RNA interference (RNAi) screen as a first step to address this question.
MATERIALS AND METHODS
Cell lines and viruses.
H1299 cells were propagated in RPMI 1640 medium containing 10 mM HEPES and 10% fetal bovine serum (FBS). H1299 cells were transfected with PXJ41neo vector encoding renilla luciferase and grown in medium containing 500 μg/ml of G418 (Sigma-Aldrich, St. Louis, MO) for 3 weeks before expansion of an antibiotic-resistant cell line (H1299-RL). Huh7 cells were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% FBS at 37°C in a 5% CO2 incubator.
The Vero cell-adapted Beaudette strain of recombinant infectious bronchitis virus luciferase (IBV-Luc), with its open reading frame (ORF) 3a-3b replaced with the firefly luciferase gene using an in vitro ligation protocol (20), was generated by infection of Vero cells with IBV-Luc at a multiplicity of infection (MOI) of approximately 0.1 in serum-free DMEM followed by 20 h of incubation. IBV stocks were prepared through lysis of infected cells with three repeated freeze-thaw cycles. Lysates were spun down at 10,000 relative centrifugal force (RCF) to remove cell debris and stored at −80°C until use. Recombinant virus expressing IBV-N fused to a Flag epitope (IBV-N-Flag) was also generated using similar protocols (21).
The HCoV-229E strain (ATCC VR-740) used in this study was directly purchased from the ATCC.
Chemical inhibitors.
MG132, bafilomycin A1 (Baf-A1), brefeldin A (BFA), and cycloheximide were purchased from Calbiochem Merck Millipore (Billerica, MA), while Golgicide A was from Sigma-Aldrich.
siRNA screen assay.
The commercial siRNA library (Dharmacon siGenome) (Thermo-Fisher Scientific, San Jose, CA) was spotted in 384-well black μClear plates (Grenier Bio-One, Frickenhausen, Germany). Using reverse transfection, siRNAs were transiently transfected at a final concentration of 25 nM per well using 0.25 μl DharmaFECT2 reagent in 7.25 μl of RPMI medium, according to the manufacturer's protocol. After 20 min of complex formation, cells were dispensed at a density of 3,000 cells per well using a Multidrop Combi dispenser (Thermo-Fisher Scientific). Medium was removed at 72 h posttransfection, and cells were washed with phosphate-buffered saline (PBS) twice before infection with IBV-Luc for 16 h. Cells were harvested with a Dual-Glo system (Promega, Madison, WI), and luminescence signals for both firefly and renilla luciferase activities were acquired with an Infinite M200 luminometer (Tecan, Durham, NC).
Data formatting, normalization, and screen quality control.
Screen quality control was performed with ScreenSifter (www.screensifter.com). Each replicate's firefly and renilla luciferase scores were normalized by z-score by plate: Z = (Xi − Xm)/σx, where Xi is the firefly signal intensity of the gene i, Xm is the mean firefly signal intensities of all the genes per plate, and σx is the standard deviation [SD] of firefly signal intensities of the genes per plate). After normalization, the Pearson correlation of firefly scores was 0.71.
Gene annotation and subcellular localization, protein networks, and comparison of screens.
Annotation information was from Gene Ontology (GO) (22), GeneCards (www.genecards.org) (23), and NCBI (http://www.ncbi.nlm.nih.gov/gene). Gene enrichment was analyzed using DAVIDS bioinformatics (http://david.abcc.ncifcrf.gov/) (24) and ScreenSifter. The protein network was created using a combination of STRING (http://string-db.org/) (25), Cytoscape, and ScreenSifter.
siRNAs.
siRNAs for primary and secondary screens were from Dharmacon's siGenome series. Pooled siRNA against Arf1, GBF1, CUL3, RBX1, and NEDD8 was from Dharmacon OnTargetPlus. One siRNA against valosin-containing protein (VCP) (sense strand, 5′-AAGUAGGGUAUGAUGACAUUG-3′; antisense strand, 5′-CAAUGUCAUCAUACCCUACUU-3′) was from Sigma-Aldrich.
Western blotting.
Anti-IBV-N, anti-IBV-S, and anti-NS3 antibodies (1:4,000) were raised as previously described (26). Mouse monoclonal antibodies against double-stranded RNA (dsRNA) were purchased from Scions, Hungary. Antibodies against β-tubulin (1:1,000), actin (1:1,000), and VCP (1:1,000) were purchased from Sigma-Aldrich, and antibodies against EEA1 (early endosomal antigen 1) (1:500), GBF1 (1:1,000), and Arf1 (1:1,000) were obtained from Abcam (Cambridge, MA).
Immunofluorescence.
Cells on coverslips were fixed with 4% formaldehyde, permeabilized with 0.2% Triton X-100, and incubated with the following antibodies: mouse anti-dsRNA (English & Scientific Consulting Bt., Hungary) (1:500), rabbit anti-Flag (Sigma-Aldrich) (1:250), mouse anti-Flag (Sigma-Aldrich) (1:250), and mouse anti-EEA1 (BD Biosciences, Franklin Lakes, NJ) (1:300).
Viral plaque assay and 50% tissue culture infective dose (TCID50) method.
Viral titers for IBV infection were determined using plaque assays. Briefly, Vero cells were seeded into 6-well plates 24 h prior to infection with 200 μl of 10-fold-serially diluted virus stock. After 1 h of absorption at 37°C, unbound virus particles were removed and cells were washed twice with PBS and maintained in 2.5 ml of DMEM containing 1% FBS and 1% carboxymethyl cellulose for 3 days. The cells were fixed with 4% paraformaldehyde and then stained with 0.1% toluidine blue. The number of plaques was counted, and the virus titer was calculated as PFU per milliliter.
Viral titers of HCoV-229E were also determined using the median tissue culture infectious dose (TCID50) per milliliter in Huh7 cells seeded in 96-well microplates.
Subcellular fractionation.
Reverse transfection of 4 million cells was performed with siRNAs at a final concentration of 30 nM per well using 160 μl DharmaFECT2 in 1.6 ml of RPMI medium. At 72 h posttransfection, cells were incubated with IBV (MOI, 0.25) for 4 h at 37°C. Infected cells were washed with ice-cold PBS, disrupted using a cell scraper in homogenization buffer (HB) (250 mM sucrose, pH 7.4, Tris, 1 mM EDTA) in the presence of protease (Complete EDTA-Free; Roche Applied Science, Indianapolis, IN) and phosphatase (PhosphoHalt; Roche Applied Science) inhibitors, and homogenized using a 22-gauge needle. Postnuclear supernatant (PNS) was obtained after centrifugation at 2,000 × g for 5 min at 4°C, and crude separation of the endosomal fractions was achieved via a sucrose flotation discontinuous gradient. The sucrose concentration of the PNS was adjusted to 40.6% by the addition of 62% sucrose (1:1.2, vol/vol), and the PNS was loaded on the bottom of an SW41 ultracentrifuge tube (Beckman, Fullerton, CA). The suspension was overlaid sequentially with 35% sucrose (2.5 ml), 25% sucrose (2 ml), and finally HB (1.5 ml). After 1 h of centrifugation at 100,000 × g at 4°C, 1-ml fractions were collected and subjected to RNA and protein extraction using TRIzol reagent (Invitrogen, Carlsbad, CA), according to the manufacturer's protocol.
RESULTS
Identification of cellular factors involved in coronavirus replication.
The infectious bronchitis virus (IBV) was used as a model coronavirus for its ease of large-scale handling and genetic manipulation. We conducted the screen using an siRNA pool library targeting 21,121 different genes in H1299 cells, a lung carcinoma cell line permissive for IBV infection (27) (Fig. 1A).
FIG 1.
Genome-wide RNAi screen for cellular factors affecting IBV replication. (A) Assay overview. H1299-RL was infected with rIBV-Luc, and 16 h later, both firefly and renilla luciferases were measured. (B) Genome-wide RNAi screen. Average Z scores of renilla and firefly luciferase activities plotted on x and y axes, respectively. A cutoff of 2.5 SD from nontargeting control siRNAs defines hits. (C) Workflow of the validation process. (D) Gene ontology analysis was performed on validated hits. Graphs show statistically significant (P < 0.05) enrichment of cellular components, biological processes, and molecular functions. (E) Pie chart of enriched cellular components. ER, endoplasmic reticulum; ECM, extracellular space.
At 72 h post-siRNA transfection, cells were infected with IBV-Luc, a recombinant strain of IBV expressing firefly luciferase (20). Firefly luminescence was measured at 16 h postinfection (hpi), which corresponds to peak firefly activity and includes some secondary infection of neighboring cells (data not shown). It should theoretically allow identification of host factors involved at any stage of infection.
To assess RNAi-associated cellular toxicity, H1299 cells were stably transfected with renilla luciferase, whose signal correlates with cell number. To eliminate false positives, samples with low cell numbers are usually excluded (28, 29, 30). However, this thresholding can lead to false negatives, genes with a real impact on viral infection but also a certain depletion-associated cytotoxicity. To optimize selection, we plotted renilla against firefly average luciferase z-scores. For most wells, there was a direct correlation between the two signals. However, this trend deviated from linearity at higher renilla luciferase values, suggesting that virus production was limited.
To confirm this viral replication saturation at high cell densities, a range of 2,500 to 12,000 cells/well was infected with IBV-Luc. Firefly signal increased linearly with cell density but saturated above ∼7,500 cells/well (data not shown). Under screen conditions, cellular density was estimated to rise from ∼3,000 to a maximum of ∼10,000 cells (>95% confluence) after 3 days of culture, thus falling within saturation range. The infection plateau was not due to a limiting amount of infectious particles, as an increasing multiplicity of infection (MOI) only marginally increased firefly luciferase activity (data not shown). Instead, a less efficient infection process due to high cell confluence is a likely explanation.
The Michaelis-Menten equation was empirically found to best account for this saturation effect in the genome-wide screen. By using this equation and setting a threshold at 2.5 standard deviations (SD) away from nontargeting controls, we identified 263 siRNA pools decreasing IBV firefly luciferase activity when silenced. On the other hand, enhancement of virus replication by 130 siRNA pools suggests that the corresponding genes may have protective roles against coronavirus infection (Fig. 1D).
Eighty-three cellular factors validated by at least two independent RNAs.
Of the 263 siRNA pools identified as supporting coronavirus infection, those found not to correspond to mRNAs in the current NCBI database (pseudogenes, RNA genes, or withdrawn mRNA records) were excluded from further analysis. The remaining 225 siRNA pools were subjected to a validation screen with each of the four siRNAs from the pool tested individually. Eighty-three (39%) of these pools had at least two out of four siRNAs that significantly (2 SD) decreased IBV replication compared to the nontargeting control and thus are unlikely to be due to off-target effects. Thus, we propose that the 83 corresponding genes are host factors required for IBV infection (Fig. 1C).
GO analysis.
Using Gene Ontology (GO), 76.2%, 70%, and 66.2% of the 83 genes were classified as involved in biological processes, cellular components, and molecular processes, respectively (Fig. 1D). A significant number of proteins localized to membranous compartments in the cell (n = 19), including those residing on the plasma membrane (n = 9), endoplasmic reticulum (ER) (n = 3), and Golgi apparatus (n = 5) (Fig. 1E), consistent with the known extensive interactions and modifications of the host membranous structures for virus entry, fusion, replication, and secretion (31, 32).
In spite of coronavirus replication and transcription occurring almost exclusively in the cytoplasm, proteins localized to the nucleus were also enriched (n = 38), with approximately one-third (n = 13) residing also in the nucleolus (Fig. 1E). This subcellular localization is due in part to genes involved in RNA splicing and transcription regulation (Fig. 1D; also see below).
Proteins in cellular processes involved in coronavirus replication are enriched in the screen results.
Among the biological processes GO, RNA splicing appeared prominently (Fig. 1D), and several other genes were associated with the spliceosomal cellular compartment GO. Together, this group of genes was largely represented among hits (Fig. 2A). The screen identified two members of the heterogenous nuclear ribonucleoprotein (hnRNP) family, hnRNP L and hnRNP N3, which have been recently identified to bind to IBV N protein. They are closely related to a known regulator of coronavirus, hnRNP A1, and form part of an interaction network that connects to coronavirus RNA and protein (NSP12) (Fig. 2B). Other members of this potential network include 10 other genes (CDC5L, CWC15, DHX8, PLRG1, PRPF18, PRPF6, SART1, SNRPD1, U2AF1, and U2AF2) that either have been connected to hnRNPs or have a proposed function in the spliceosome (Fig. 2B).
FIG 2.
Common cellular processes important in coronavirus replication. (A) RNA splicing factors and spliceosomal components among hits. (B) RNA splicing genes and viral RNA (TRS, transcriptional regulatory sequence) interaction network. (C) Interaction network between CoV-interacting factors and hits. UTR, untranslated region. (D) Membrane-associated genes present in the screen. (E) Interactions between ubiquitin-proteasome pathway hits and coronavirus-interacting partners.
Another host process regulated in response to viral infection is protein synthesis. The eukaryotic translation initiation factor 3, subunit E (eIF3f), identified in our screen, interacts with six other known coronavirus-interacting proteins (Fig. 2C). Among them are two other members of the eIF3 family that had been previously identified to be involved in SARS-CoV replication and to be important for modulating host translation of inhibitory cytokines and chemokines (18, 33). Two other factors identified, PES1 and RSL24D1, also interact with eIF3E and with factors otherwise known to interact with viral proteins (Fig. 2C).
Membrane trafficking genes.
Several hits were linked to a cellular compartment GO linked to membranes and were known regulators of membrane traffic processes (Fig. 2D). Among them are Vps4B, also known as the charged multivesicular protein 4b (CHMP4B), which is a key component of the ESCRT complex that regulates multivesicular body sorting, and the Golgi apparatus-associated brefeldin A (BFA)-resistant GEF 1 (GBF1), which has been shown to be required for multiple viruses (34, 35). GBF1 is a regulator of ADP-ribosylation factors such as ADP-ribosylation factor 1 (Arf1) (36). Despite not being a direct “hit” in our screen, Arf1 scored close to the threshold. Interestingly, the related Arf5 scored high in our screen and was recently identified as required for dengue virus (37). In a screen targeting the druggable genome library, Burkard et al. identified several proteins mediating late endosomal maturation as being important for mouse hepatitis virus (MHV) infection (38). These proteins, however, were dispensable for IBV infection (not shown), reflecting the fact that different endocytotic processes are probably required by specific members of the coronavirus family.
The UPS is highly represented.
The interaction network established above identified seven factors associated with the ubiquitin-proteasome system (UPS), interacting extensively among themselves as well as with other known CoV-interacting proteins (Fig. 2E). In particular, six known CoV-interacting proteins interact with the Nedd8 protein, and our screen identified three other Nedd8-interacting proteins. Two of them, Cullin-3 (Cul3) and ring box 1 (Rbx1), form the core of the BTB-CUL3-RBX1 E3 ligase complex (39). Activation of this complex requires covalent attachment of Nedd8 by ube2m, a ubiquitin-conjugating enzyme also identified in our screen (40).
The requirement for Cul3, Rbx1, and Nedd8 was confirmed with an independent secondary assay using Western blot analysis against viral protein at 16 hpi but also 8 hpi. A reduction in viral protein expression is associated with knockdowns of these genes at both time points, suggesting a requirement for these proteins during early infection stages (data not shown). It is not clear yet what the relevant ubiquitinated substrates are, but it is noticeable that depletion of PSMD14 and other proteasome subunits had a strong inhibitory effect on viral infection. This proteasome requirement is also consistent with previous findings on the inhibitory effect of proteasome inhibitors on CoV replication (41, 42).
Our screen results also implicate two other UPS-related proteins: valosin-containing protein (VCP/p97) and ubiquitin fusion degradation protein 1-like protein (Ufd1L) (Fig. 2E). The ternary complex of Vcp-Ufd1L-Npl4 is one of the key mediators of ER-associated degradation process (ERAD) (43, 44). All three components were identified as primary hits in our screen, although only VCP and Ufd1L could be validated in the deconvoluted screen.
As depletion of VCP induced one of the strongest inhibitory effects on IBV replication and had not been identified as a host factor for coronavirus infection before, we chose to further characterize its role in coronavirus infection.
We first compared multiple anti-VCP siRNAs. While the screen siRNA pool (siVCP-Pool) reduced IBV-Luc activity by 14.4-fold, the 4 individual siRNAs in the pool (siVCP-05, -06, -07, and -08) reduced IBV-Luc activity by 13-, 10-, 4-, and 6-fold, respectively. In addition, another custom-designed siRNA (siVCP-Sig) resulted in a 23-fold reduction (data not shown). This siRNA was used in subsequent experiments.
VCP is required for human coronavirus infection.
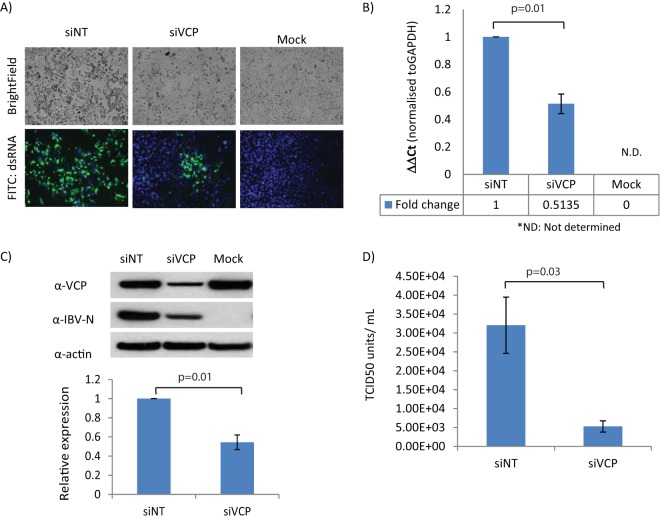
To investigate if VCP is required by coronaviruses in general, the requirement for VCP in human coronavirus 229E (HCoV-229E) infection was determined. HCoV-229E infection of susceptible Huh7 cells resulted in more extensive cytopathic effects and was observed in susceptible Huh7 cells treated with control siRNA rather than those with VCP silenced. In addition, when these cells were stained for the presence of dsRNA, the levels of dsRNA detected were significantly lower in VCP-knockdown cells than in control (Fig. 3A). The lower rate of viral replication associated with VCP depletion was also confirmed with quantitative real-time PCR using primers specific to HCoV-229E genomic RNA. Viral genomic RNA detected in cells treated with siVCP was 2-fold lower than that in controls (Fig. 3B). Antibodies against IBV nucleocapsid protein (IBV-N), which cross-react with HCoV-229E nucleocapsid protein (N), were also used to determine expression of viral protein in the absence of VCP. In these experiments, depletion of VCP with RNAi consistently resulted in a decrease in HCoV-N protein expression (Fig. 3C).
FIG 3.
Depletion in VCP leads to decreased human coronavirus 229E (HCoV-229E) replication. (A) Huh7 cells transfected with nontargeting siRNA (siNT) or VCP siRNA (siVCP) were observed for cytopathic effect under a light microscope after 48 h of HCoV-229E infection (top panel). Cells were also then fixed, infected, and stained for dsRNA expression and nuclei by Hoechst staining (bottom panel). FITC, fluorescein isothiocyanate. (B) Viral genomic RNA (+gRNA) levels were then analyzed using quantitative real-time PCR. Error bars represent standard deviations obtained from two independent experiments performed in triplicate. (C) The level of nucleocapsid (N) expression in HCoV-229E-infected cells was probed using cross-reactive antibodies raised against IBV-N. (D) The titer of viral particles released into the respective culture supernatant was first determined with the method of TCID50 per milliliter and then confirmed via standard plaque assay.
Finally, the titers of viral particles released in culture supernatant were determined by the method of median tissue culture infectious dose (TCID50) per milliliter. Consistent with all the previous results, the titer of viral particles released from siVCP-treated cells was significantly lower than that in control (Fig. 3D). The reduction in viral titer was confirmed independently with standard viral plaque assay (Fig. 3D). Collectively, the results suggest that VCP may be required for coronavirus in general.
VCP is involved in early stages of infection.
To examine at which stage VCP is required, IBV-infected cells were harvested at 10 hpi, when there is minimal secretion of virus and secondary infection. Silencing of VCP reduced firefly luciferase activity by nearly 8-fold compared with cells treated with nontargeting siRNA control (Fig. 4A), suggesting a requirement in the initial phase of infection.
FIG 4.
Role of valosin-containing protein (VCP) in early virus replication. (A) Effect of VCP depletion on infectious bronchitis virus (IBV)-Luc replication at 10 or 16 hpi. Error bars represent SDs from average luciferase activity from two independent experiments with duplicates. (B) Effect of VCP depletion on dsRNA production, measured at 8 hpi with a specific monoclonal antibody. Cells were also stained with anti-IBV-S antibody, and nuclei were stained with Hoechst stain. DAPI, 4′,6-diamidino-2-phenylindole. (C) Effect of VCP knockdown on IBV-N expression at 8 and 16 hpi as measured by Western blotting. (D and E) Role of VCP in virion attachment. Attached viral particles were measured by counting the number of CFU by standard plaque assay (D) or via the detection of genomic RNA (+gRNA) levels by RT-PCR (E). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was measured as a loading control. (F and G) Role of VCP in virus entry. Internalized virus particles were quantified by plaque assay (F) as in panel D or by RT-PCR (G). NT, nontargeting.
The production of double-stranded RNA (dsRNA) intermediates, a hallmark of viral transcription/replication, at 10 hpi was evaluated by immunofluorescence using a dsRNA-specific monoclonal antibody, revealing that silencing of VCP significantly reduced dsRNA levels (Fig. 4B). These results are consistent with a concomitant reduction in viral protein expression, as reduced levels of IBV-S and IBV-N proteins, revealed by immunofluorescence and Western blot analysis, respectively (Fig. 4B and C), were also associated with VCP depletion. Furthermore, syncytium formation, as revealed by the IBV-S antibody, was evident in nontargeting siRNA (siNT)-treated control cells but not observed in cells depleted of VCP (Fig. 4B).
VCP is not required for viral attachment to cell surface and virus entry.
To examine virion attachment to cell surface, cells treated with either control siNT or siVCP were incubated with IBV-Luc at an MOI of approximately 0.5 for 1 h at 4°C to allow for virus adsorption. Unbound virus particles were removed by washing, and bound viruses were quantified using standard plaque assay and did not reveal any decrease (Fig. 4D). Similarly, quantification of bound virus using reverse transcription-PCR (RT-PCR) did not result in a decreased amount of virus-positive genomic RNA detected on the surface of the infected VCP-depleted cells (Fig. 4E), suggesting that VCP is not required for the virions to bind to the host cell.
Next, we tested virus internalization by incubating cells first with IBV-Luc at an MOI of approximately 0.5 for 1 h at 4°C before washing and temperature shift to 37°C for an additional 1 h to allow synchronized entry. Proteinase K was then added to remove uninternalized virus before harvesting infected cells by freeze-thawing. Internalized virions were quantified by both titration plaque assay (Fig. 4F) and RT-PCR (Fig. 4G), and no significant reduction was detected in VCP-depleted cells, suggesting that VCP is also not required for virus entry.
Silencing of VCP inhibits early-stage degradation of viral N protein.
While monitoring the amount of the viral protein N at early time points, we observed higher levels of N protein in VCP-depleted cells at 4 hpi (Fig. 5A). This effect is transient, as by 8 hpi, N protein levels in control cells were dramatically increased over those at 4 hpi and became significantly higher than those in VCP-depleted cells (Fig. 5A), where N protein remained constant, which is consistent with the block in the infectious process detected with other methods.
FIG 5.
Role of valosin-containing protein (VCP) in early-stage degradation of N protein. (A) Representative blot of IBV-N in VCP-depleted or control-treated (NT) cells infected with IBV for 4 or 8 h. Intensities of the bands were quantified and represented as an average of data from two independent experiments. (B) Comparison of effects of VCP and GBF1 depletion on IBV-N levels. (C) Effect of cycloheximide (CHX; 100 μg/ml) on IBV-N accumulation. β-Tubulin was used as a loading control. sgRNA, subgenomic RNA. (D) Effect of DMSO, MG132 (10 μM), brefeldin A (BFA; 5 μg/ml), Golgicide A (GCA; 10 μg/ml), or bafilomycin A1 (Baf-A1; 1 μM) applied 2 h prior to infection with IBV (MOI, 0.5). The cell lysate was probed for IBV-N as well as actin as a loading control.
To determine if the difference at 4 hpi could be observed for other genes, we tested siGBF1 but did not find similar elevated N protein levels (Fig. 5B).
Typically, at 4 hpi, only a minimal amount of viral RNA and protein can be detected (data not shown). We therefore hypothesized that the N protein detected corresponds to the virion-associated rather than the neosynthesized pool. Consistent with this hypothesis, in cells pretreated with 10 μg/ml of cycloheximide, a protein translation inhibitor, levels of N protein were not affected in control or siVCP-treated cells (Fig. 5C). In contrast, at 8 h, these levels were dramatically reduced in control cells, indicating that most of the N protein detected at this time point is neosynthesized. Interestingly, in VCP-depleted cells, N protein levels were still not affected at 8 h, indicating that the IBV-N detected there came mostly from the infecting virions.
These data suggest that IBV-N associated with infecting viruses is degraded at early stages of infection. Interestingly, cells pretreated with the proteasome inhibitor MG132 for 2 h prior to infection display significantly increased N protein levels at 4 hpi compared with control and dimethyl sulfoxide (DMSO)-treated cells (Fig. 5D). Treatment with either 5 μg/ml BFA or 10 μM Golgicide A1 did not result in increased N levels, consistent with the GBF1 depletion experiment results. On the other hand, treatment with bafilomycin A1 (Baf-A1), which targets proton pump ATPases, was comparable to MG132 in effect (Fig. 5D).
Together, our results suggest that the N protein associated with infecting virions is degraded before a new pool is neosynthesized and that this process requires endosomal acidification and proteasome function.
Silencing of VCP results in accumulation of virus in early endosomes.
Since VCP has been linked to the ubiquitin-proteasome system, a possible explanation for the accumulation of virion-associated N protein is that VCP is directly required for its degradation. In this model, VCP would act after the release of the viral nucleoprotein from the endosomal system. However, during the course of this study, VCP was reported to be implicated in endosomal maturation after VCP depletion (45, 46) and could therefore result in a trapping of viral particles in endosomes before cytosolic release.
To distinguish between these possibilities, the presence of intracellular viral particles was tested by immunofluorescence. Since the IBV-N antibody is incompatible with immunofluorescence, cells were infected with another recombinant virus expressing IBV-N fused to a Flag epitope (IBV-N-Flag) (21). The number of Flag-positive vesicular structures was counted in 10 random cells from each treatment group and compared with the control group, demonstrating that siVCP-treated cells contained more IBV-N-positive vesicles (data not shown).
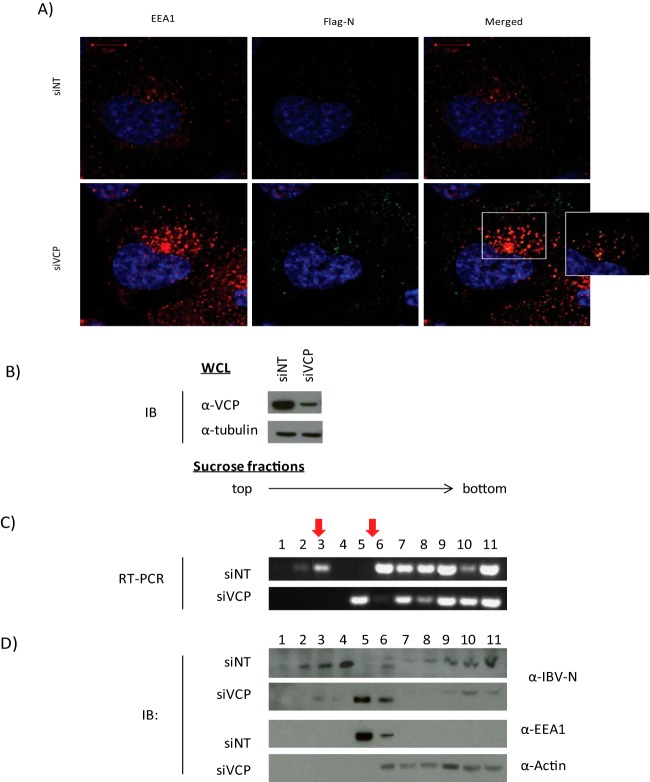
Infected cells were also costained for the early endosomal antigen 1 (EEA1), and numerous and enlarged vesicles positive for EEA1 were observed in siVCP-treated cells (Fig. 6A). Anti-Flag staining revealed that virion-associated IBV-N protein colocalized extensively with these enlarged endosome clusters (Fig. 6A), suggesting that virion particles accumulate in an endosomal compartment in the absence of VCP.
FIG 6.
Valosin-containing protein (VCP) depletion leads to accumulation of viral particles in early endosome vesicles. (A) H1299 cells transfected with nontargeting or anti-VCP siRNA (siNT or siVCP, respectively) were infected with recombinant IBV expressing Flag-tagged N protein (rIBV-FlagN) for 4 h, fixed, permeabilized, and stained for EEA1 (tetramethyl rhodamine isocyanate [TRITC], red) or anti-Flag (fluorescein isothiocyanate [FITC], green). A magnified view of colocalized particles is provided in the square inset. (B to D) Subcellular fractionation of IBV-N in VCP-silenced or siNT-treated cells using step sucrose gradient flotation. (B) Western blot analysis of whole-cell lysate (WCL). (C) RT-PCR against viral genomic RNA. (D) Western blot analysis of sucrose fractions against IBV-N, IBV-S, EEA1, and actin. Results are representative of three independent experiments. Tubulin and actin were used as loading controls.
IBV-infected cells were next subjected to subcellular fractionation using a sucrose flotation step and discontinuous gradient 4 h after infection. VCP depletion was verified by Western blotting (Fig. 6B). Using a modified protocol first described by Gruenberg and Gorvel for endosomal fractionation (47), late and early endosomes were concentrated in the 8%/25% (fractions 2 and 3) and 25%/35% (fractions 5 and 6) interphases, respectively. TRIzol extraction was used to obtain both RNA and protein to detect the presence of virus particles via RT-PCR or Western blot analysis, respectively.
In both control cells and cells treated with siVCP, most of the virus genomic RNA was detected in heavier fractions (fractions 7 to 11). RNA detected from these fractions is likely to come from viral material present in the cytosol as well as that released during cell lysis. Viral RNA was also detected in fractions 2 and 3 (late endosome fractions) in the control group. In contrast, in samples where VCP was silenced, viral RNA was nearly undetectable in the late endosome fractions (Fig. 6C). Instead, viral RNA was observed to accumulate in early endosomes (fraction 5) together with EEA1 (Fig. 6D). Analysis of the viral protein yielded similar results. When probed with antibodies against N protein, knocking down VCP resulted in viral proteins accumulating in fractions that were also enriched with EEA1 (Fig. 6C and D). These results suggest that, in the absence of VCP, the infecting virus accumulates in an early endosomal compartment.
DISCUSSION
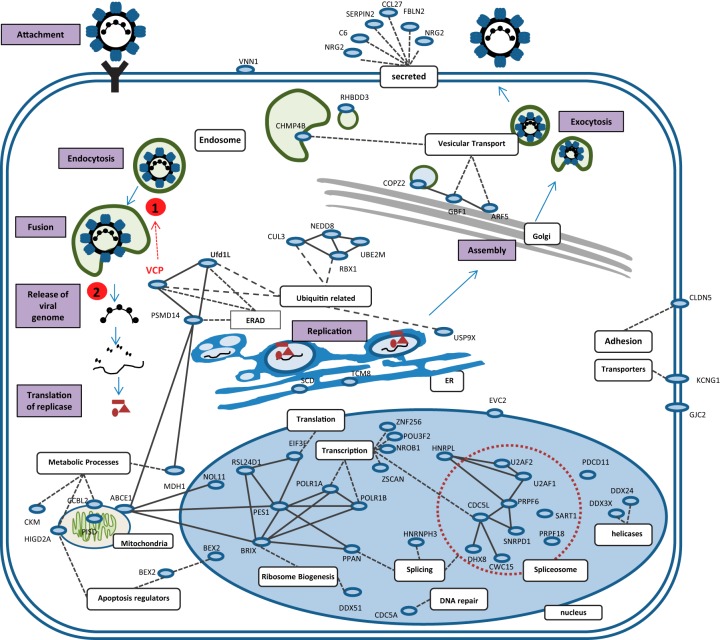
Like all viruses, CoV are highly dependent on their host cellular machinery. Until now, relatively few human genes had been functionally implicated in CoV replication. Through this genome-wide RNAi screen against IBV, we have identified 83 new cellular cofactors. Albeit IBV is an avian virus, coronaviruses are known to be able to jump species, and as we used a human lung cell line for our screen, the host factors identified are likely to be conserved across species and, to some extent, across different strains of coronaviruses. In fact, 30 hits are directly or indirectly interacting with proteins that were found to interact with SARS-CoV proteins (data not shown), supporting the notion of a relatively well conserved set of interactions. To summarize our observations, a cellular map illustrating the predominant subcellular locations and functions of hits with a presumed function is presented in Fig. 7.
FIG 7.
Cellular map illustrating predominant subcellular localization and presumed functions of hits. We proposed that depletion of VCP prevents virus particles from exiting endosomes (1) and hence inhibits the subsequent uncoating and N protein degradation event (2).
Although we did not focus our study on it, the host negative regulators of CoV promise to help us understand how cells can resist CoV infection. In fact, the ATP-binding cassette subfamily E member 1 (ABCE1) gene was an important and confirmed hit. The encoded protein has been proposed to bind to and inhibit RNase L. Activation of RNase L is part of the 2′,5′-oligoadenylate synthetase (OAS)–RNase L pathway, which is activated in response to interferon and results in a block of protein synthesis and the promotion of viral RNA degradation (48). It would be interesting to test whether ABCE1 is constitutively active in H1299 cells or whether it might be activated by CoV infection.
Another group of very interesting genes is from the RNA binding family. Indeed, a major feature of the CoV life cycle is the discontinuous transcription of its genomic RNA. Our screen results suggest that the host splicing machinery is heavily recruited to perform this task. For example, one of the hits is hnRNP L, a protein known to regulate mRNA splicing (49). Among the most interesting prospects for the development of new therapeutic approaches is the targeting of host genes required for multiple viruses. The VCP/Ufd1L complex could represent such a target, as it appears essential for CoV, dengue virus (data not shown), and West Nile virus (WNV) (shown for Ufd1L) (29). In addition, VCP was recently proposed to be required for poliovirus infection and hepatitis B (50, 51).
VCP is an abundant protein implicated in an increasing number of biological processes, including membrane fusion after mitosis (52), retrotranslocation of unfolded proteins from the endoplasmic reticulum (ER) (43, 53), spindle assembly (54), and, more recently, endosome trafficking (45, 55). Thus, VCP might be required at multiple stages of viral replication or differently, depending on the viruses. For instance, VCP was proposed to be required after viral protein replication for poliovirus (50). For IBV, VCP is required at an early step, preceding the production of viral protein and RNA replication.
VCP depletion revealed a previously uncharacterized transient degradation of the virion-associated N protein. N protein forms a tight complex with the viral RNA genome (56); it is thus possible that this degradation is required for the cytosolic release of the viral genome and productive infection. N protein is localized inside the viral envelope; thus, its degradation by the proteasome must follow the fusion of the viral envelope. It is not known, however, how neosynthesized N protein is protected from degradation. The exact mechanism by which VCP facilitates N protein degradation is also currently unclear. Hauler et al. (57) had previously reported that VCP is required for the proteasomal degradation of antibody-bound adenovirus capsid protein during the process of intracellular neutralization. This process appears to be substrate specific and may involve ATP-hydrolysis-driven unfolding and disassembly of the large capsid protein to “fit” into the proteasome core for degradation. VCP-dependent degradation of N protein may occur in a similar fashion.
The subcellular fractionation of infected cells upon VCP depletion resulted in the association of viral particles with endosomal membranes that cosegregated with EEA1. An interpretation of this result is that VCP is required for the maturation of endosomes. A block in endosomal maturation could result in a loss of acidification of the virus-containing membranes and a failure of the viral envelope to fuse with the endosomal membrane. Indeed, it has been reported elsewhere that CoV spike protein requires an acidic pH to undergo the protease cleavage that leads to fusion (32, 58).
The function of VCP in endosomal maturation is still unclear. A recent report proposes a function for VCP in membrane fusion and early endosome maturation through the segregation of EEA1 polymers (45). Another report implicates VCP in the extraction and translocation of the polyubiquitinated mannose receptor from endosomes to the cytosol (59). A complex of VCP and UBXD1 has also been shown recently to regulate endolysosomal sorting of ubiquitylated caveolin-1 (55), where the authors observed an enlargement of late endosomes/lysosomes and an impairment of multivesicular body formation. This phenotype of enlarged late endosomes/lysosomes is similar to the effect of loss of function of Cullin-3 (60). We also identified Cullin-3 in our screen. In the absence of this protein, influenza virus particles have been reported to accumulate in late endosomes/lysosomes and to exhibit defective uncoating (60). Cullin-3-depleted cells also display an accumulation of morphologically abnormal acidic late endosomes/lysosomes (LE/LY). Interestingly, Cullin ring ligases have been shown previously to function with VCP (61). However, whether Cullin-3 and VCP function together to permit CoV infection remains unclear.
More work is obviously required to sort out the precise function(s) of VCP in the endosomal system and the requirements of VCP for infection by IBV and other viruses. These efforts could be rewarding, as blocking viral release into the cytosol is a promising therapeutic strategy. Conversely, IBV-N protein degradation at the early stages of infection could represent an interesting assay to study endosomal maturation.
ACKNOWLEDGMENTS
This work was supported by the Institute of Molecular and Cell Biology (IMCB) and the Agency for Science Technology and Research (A*STAR), Singapore.
We also thank Ahmed Sayadi for his help with the protein interaction illustration.
REFERENCES
- 1.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. 2012. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med 367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 2.Peiris JS, Guan Y, Yuen KY. 2004. Severe acute respiratory syndrome. Nat Med 10:S88–S97. doi: 10.1038/nm1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van der Hoek L. 2007. Human coronaviruses: what do they cause? Antivir Ther 12:651–658. [PubMed] [Google Scholar]
- 4.Saif LJ. 2010. Bovine respiratory coronavirus. Vet Clin North Am Food Anim Pract 26:349–364. doi: 10.1016/j.cvfa.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laude H, Van Reeth K, Pensaert M. 1993. Porcine respiratory coronavirus: molecular features and virus-host interactions. Vet Res 24:125–150. [PubMed] [Google Scholar]
- 6.Cavanagh D. 2007. Coronavirus avian infectious bronchitis virus. Vet Res 38:281–297. doi: 10.1051/vetres:2006055. [DOI] [PubMed] [Google Scholar]
- 7.Vijgen L, Keyaerts E, Moes E, Thoelen I, Wollants E, Lemey P, Vandamme AM, Van Ranst M. 2005. Complete genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. J Virol 79:1595–1604. doi: 10.1128/JVI.79.3.1595-1604.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li W, Shi Z, Yu M, Ren W, Smith C, Epstein JH, Wang H, Crameri G, Hu Z, Zhang H, Zhang J, McEachern J, Field H, Daszak P, Eaton BT, Zhang S, Wang LF. 2005. Bats are natural reservoirs of SARS-like coronaviruses. Science 310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 9.van Boheemen S, de Graaf M, Lauber C, Bestebroer TM, Raj VS, Zaki AM, Osterhaus AD, Haagmans BL, Gorbalenya AE, Snijder EJ, Fouchier RA. 2012. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. mBio 3(6):e00473-12. doi: 10.1128/mBio.00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Breban R, Riou J, Fontanet A. 2013. Interhuman transmissibility of Middle East respiratory syndrome coronavirus: estimation of pandemic risk. Lancet 382:694–699. doi: 10.1016/S0140-6736(13)61492-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lim PL, Lee TH, Rowe EK. 2013. Middle East respiratory syndrome coronavirus (MERS CoV): update 2013. Curr Infect Dis Rep 15:295–298. doi: 10.1007/s11908-013-0344-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perlman S, Netland J. 2009. Coronaviruses post-SARS: update on replication and pathogenesis. Nat Rev Microbiol 7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Narayanan K, Huang C, Makino S. 2008. SARS coronavirus accessory proteins. Virus Res 133:113–121. doi: 10.1016/j.virusres.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oostra M, te Lintelo EG, Deijs M, Verheije MH, Rottier PJ, de Haan CA. 2007. Localization and membrane topology of coronavirus nonstructural protein 4: involvement of the early secretory pathway in replication. J Virol 81:12323–12336. doi: 10.1128/JVI.01506-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Verheije MH, Raaben M, Mari M, Te Lintelo EG, Reggiori F, van Kuppeveld FJ, Rottier PJ, de Haan CA. 2008. Mouse hepatitis coronavirus RNA replication depends on GBF1-mediated ARF1 activation. PLoS Pathog 4:e1000088. doi: 10.1371/journal.ppat.1000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu LH, Huang M, Fang SG, Liu DX. 2011. Coronavirus infection induces DNA replication stress partly through interaction of its nonstructural protein 13 with the p125 subunit of DNA polymerase delta. J Biol Chem 286:39546–39559. doi: 10.1074/jbc.M111.242206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu L, Khadijah S, Fang S, Wang L, Tay FP, Liu DX. 2010. The cellular RNA helicase DDX1 interacts with coronavirus nonstructural protein 14 and enhances viral replication. J Virol 84:8571–8583. doi: 10.1128/JVI.00392-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pfefferle S, Schopf J, Kogl M, Friedel CC, Muller MA, Carbajo-Lozoya J, Stellberger T, von Dall'Armi E, Herzog P, Kallies S, Niemeyer D, Ditt V, Kuri T, Züst R, Pumpor K, Hilgenfeld R, Schwarz F, Zimmer R, Steffen I, Weber F, Thiel V, Herrler G, Thiel HJ, Schwegmann-Wessels C, Pöhlmann S, Haas J, Drosten C, von Brunn A. 2011. The SARS-coronavirus-host interactome: identification of cyclophilins as target for pan-coronavirus inhibitors. PLoS Pathog 7:e1002331. doi: 10.1371/journal.ppat.1002331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cruz JL, Sola I, Becares M, Alberca B, Plana J, Enjuanes L, Zuñiga S. 2011. Coronavirus gene 7 counteracts host defenses and modulates virus virulence. PLoS Pathog 7:e1002090. doi: 10.1371/journal.ppat.1002090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen H, Fang SG, Chen B, Chen G, Tay FP, Liu DX. 2009. Towards construction of viral vectors based on avian coronavirus infectious bronchitis virus for gene delivery and vaccine development. J Virol Methods 160:48–56. doi: 10.1016/j.jviromet.2009.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fang S, Xu L, Huang M, Qisheng Li F, Liu DX. 2013. Identification of two ATR-dependent phosphorylation sites on coronavirus nucleocapsid protein with nonessential functions in viral replication and infectivity in cultured cells. Virology 444:225–232. doi: 10.1016/j.virol.2013.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium Nat Genet 25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rebhan M, Chalifa-Caspi V, Prilusky J, Lancet D. 1998. GeneCards: a novel functional genomics compendium with automated data mining and query reformulation support. Bioinformatics 14:656–664. doi: 10.1093/bioinformatics/14.8.656. [DOI] [PubMed] [Google Scholar]
- 24.Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 25.Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, Bork P, von Mering C. 2009. STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res 37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nasirudeen AM, Wong HH, Thien P, Xu S, Lam KP, Liu DX. 2011. RIG-I, MDA5 and TLR3 synergistically play an important role in restriction of dengue virus infection. PLoS Negl Trop Dis 5:e926. doi: 10.1371/journal.pntd.0000926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tay FP, Huang M, Wang L, Yamada Y, Liu DX. 2012. Characterization of cellular furin content as a potential factor determining the susceptibility of cultured human and animal cells to coronavirus infectious bronchitis virus infection. Virology 433:421–430. doi: 10.1016/j.virol.2012.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sessions OM, Barrows NJ, Souza-Neto JA, Robinson TJ, Hershey CL, Rodgers MA, Ramirez JL, Dimopoulos G, Yang PL, Pearson JL, Garcia-Blanco MA. 2009. Discovery of insect and human dengue virus host factors. Nature 458:1047–1050. doi: 10.1038/nature07967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krishnan MN, Ng A, Sukumaran B, Gilfoy FD, Uchil PD, Sultana H, Brass AL, Adametz R, Tsui M, Qian F, Montgomery RR, Lev S, Mason PW, Koski RA, Elledge SJ, Xavier RJ, Agaisse H, Fikrig E. 2008. RNA interference screen for human genes associated with West Nile virus infection. Nature 455:242–245. doi: 10.1038/nature07207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tai AW, Benita Y, Peng LF, Kim SS, Sakamoto N, Xavier RJ, Chung RT. 2009. A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host Microbe 5:298–307. doi: 10.1016/j.chom.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Snijder EJ, van der Meer Y, Zevenhoven-Dobbe J, Onderwater JJ, van der Meulen J, Koerten HK, Mommaas AM. 2006. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J Virol 80:5927–5940. doi: 10.1128/JVI.02501-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Belouzard S, Millet JK, Licitra BN, Whittaker GR. 2012. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses 4:1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiao H, Xu LH, Yamada Y, Liu DX. 2008. Coronavirus spike protein inhibits host cell translation by interaction with eIF3f. PLoS One 3:e1494. doi: 10.1371/journal.pone.0001494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hsu NY, Ilnytska O, Belov G, Santiana M, Chen YH, Takvorian PM, Pau C, van der Schaar H, Kaushik-Basu N, Balla T, Cameron CE, Ehrenfeld E, van Kuppeveld FJ, Altan-Bonnet N. 2010. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell 141:799–811. doi: 10.1016/j.cell.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goueslain L, Alsaleh K, Horellou P, Roingeard P, Descamps V, Duverlie G, Ciczora Y, Wychowski C, Dubuisson J, Rouillé Y. 2010. Identification of GBF1 as a cellular factor required for hepatitis C virus RNA replication. J Virol 84:773–787. doi: 10.1128/JVI.01190-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Casanova JE. 2007. Regulation of Arf activation: the Sec7 family of guanine nucleotide exchange factors. Traffic 8:1476–1485. doi: 10.1111/j.1600-0854.2007.00634.x. [DOI] [PubMed] [Google Scholar]
- 37.Kudelko M, Brault JB, Kwok K, Li MY, Pardigon N, Peiris JS, Bruzzone R, Desprès P, Nal B, Wang PG. 2012. Class II ADP-ribosylation factors are required for efficient secretion of dengue viruses. J Biol Chem 287:767–777. doi: 10.1074/jbc.M111.270579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burkard C, Verheije MH, Wicht O, van Kasteren SI, van Kuppeveld FJ, Haagmans BL, Pelkmans L, Rottier PJ, Bosch BJ, de Haan CA. 2014. Coronavirus cell entry occurs through the endo-/lysosomal pathway in a proteolysis-dependent manner. PLoS Pathog 10:e1004502. doi: 10.1371/journal.ppat.1004502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pintard L, Willems A, Peter M. 2004. Cullin-based ubiquitin ligases: Cul3-BTB complexes join the family. EMBO J 23:1681–1687. doi: 10.1038/sj.emboj.7600186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Plafker KS, Singer JD, Plafker SM. 2009. The ubiquitin conjugating enzyme, UbcM2, engages in novel interactions with components of cullin-3 based E3 ligases. Biochemistry 48:3527–3537. doi: 10.1021/bi801971m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raaben M, Posthuma CC, Verheije MH, te Lintelo EG, Kikkert M, Drijfhout JW, Snijder EJ, Rottier PJ, de Haan CA. 2010. The ubiquitin-proteasome system plays an important role during various stages of the coronavirus infection cycle. J Virol 84:7869–7879. doi: 10.1128/JVI.00485-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu GY, Lai MM. 2005. The ubiquitin-proteasome system facilitates the transfer of murine coronavirus from endosome to cytoplasm during virus entry. J Virol 79:644–648. doi: 10.1128/JVI.79.1.644-648.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wolf DH, Stolz A. 2012. The Cdc48 machine in endoplasmic reticulum associated protein degradation. Biochim Biophys Acta 1823:117–124. doi: 10.1016/j.bbamcr.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 44.Ye Y, Meyer HH, Rapoport TA. 2001. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature 414:652–656. doi: 10.1038/414652a. [DOI] [PubMed] [Google Scholar]
- 45.Ramanathan HN, Ye Y. 2012. The p97 ATPase associates with EEA1 to regulate the size of early endosomes. Cell Res 22:346–359. doi: 10.1038/cr.2011.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meyer H, Bug M, Bremer S. 2012. Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat Cell Biol 14:117–123. doi: 10.1038/ncb2407. [DOI] [PubMed] [Google Scholar]
- 47.Gruenberg J, Gorvel JP. 1997. In vitro reconstitution of endocytic vesicle fusion, p 186 In Graham JM, Rickwood D (ed), Subcellular fractionation: a practical approach. IRL Press, New York, NY. [Google Scholar]
- 48.Silverman RH. 2007. Viral encounters with 2′,5′-oligoadenylate synthetase and RNase L during the interferon antiviral response. J Virol 81:12720–12729. doi: 10.1128/JVI.01471-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heiner M, Hui J, Schreiner S, Hung LH, Bindereif A. 2010. HnRNP L-mediated regulation of mammalian alternative splicing by interference with splice site recognition. RNA Biol 7:56–64. doi: 10.4161/rna.7.1.10402. [DOI] [PubMed] [Google Scholar]
- 50.Arita M, Wakita T, Shimizu H. 2012. Valosin-containing protein (VCP/p97) is required for poliovirus replication and is involved in cellular protein secretion pathway in poliovirus infection. J Virol 86:5541–5553. doi: 10.1128/JVI.00114-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiao BY, Lin WS, She FF, Chen WN, Lin X. 2011. Hepatitis B virus X protein enhances activation of nuclear factor kappaB through interaction with valosin-containing protein. Arch Virol 156:2015–2012. doi: 10.1007/s00705-011-1099-4. [DOI] [PubMed] [Google Scholar]
- 52.Uchiyama K, Kondo H. 2005. p97/p47-mediated biogenesis of Golgi and ER. J Biochem 137:115–119. doi: 10.1093/jb/mvi028. [DOI] [PubMed] [Google Scholar]
- 53.Ye Y. 2006. Diverse functions with a common regulator: ubiquitin takes command of an AAA ATPase. J Struct Biol 156:29–40. doi: 10.1016/j.jsb.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 54.Cao K, Nakajima R, Meyer HH, Zheng Y. 2003. The AAA-ATPase Cdc48/p97 regulates spindle disassembly at the end of mitosis. Cell 115:355–367. doi: 10.1016/S0092-8674(03)00815-8. [DOI] [PubMed] [Google Scholar]
- 55.Ritz D, Vuk M, Kirchner P, Bug M, Schutz S, Hayer A, Bremer S, Lusk C, Baloh RH, Lee H, Glatter T, Gstaiger M, Aebersold R, Weihl CC, Meyer H. 2011. Endolysosomal sorting of ubiquitylated caveolin-1 is regulated by VCP and UBXD1 and impaired by VCP disease mutations. Nat Cell Biol 13:1116–1123. doi: 10.1038/ncb2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Laude H, Masters PS. 1995. The coronavirus nucleocapsid protein, p 141–158. In Siddell SG. (ed), The Coronaviridae. Plenum Press, New York, NY. [Google Scholar]
- 57.Hauler FMD, McEwan WA, Bidgood SR, James LC. 2012. AAA ATPase p97/VCP is essential for TRIM21-mediated virus neutralization. Proc Natl Acad Sci U S A 109:19733–19738. doi: 10.1073/pnas.1210659109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chu VC, McElroy LJ, Chu V, Bauman BE, Whittaker GR. 2006. The avian coronavirus infectious bronchitis virus undergoes direct low-pH-dependent fusion activation during entry into host cells. J Virol 80:3180–3188. doi: 10.1128/JVI.80.7.3180-3188.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zehner M, Chasan AI, Schuette V, Embgenbroich M, Quast T, Kolanus W, Burgdorf S. 2011. Mannose receptor polyubiquitination regulates endosomal recruitment of p97 and cytosolic antigen translocation for cross-presentation. Proc Natl Acad Sci U S A 108:9933–9938. doi: 10.1073/pnas.1102397108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huotari J, Meyer-Schaller N, Hubner M, Stauffer S, Katheder N, Horvath P, Mancini R, Helenius A, Peter M. 2012. Cullin-3 regulates late endosome maturation. Proc Natl Acad Sci U S A 109:823–828. doi: 10.1073/pnas.1118744109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.den Besten W, Verma R, Kleiger G, Oania RS, Deshaies RJ. 2012. NEDD8 links cullin-RING ubiquitin ligase function to the p97 pathway. Nat Struct Mol Biol 19:511–516. doi: 10.1038/nsmb.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]