Figure 4.
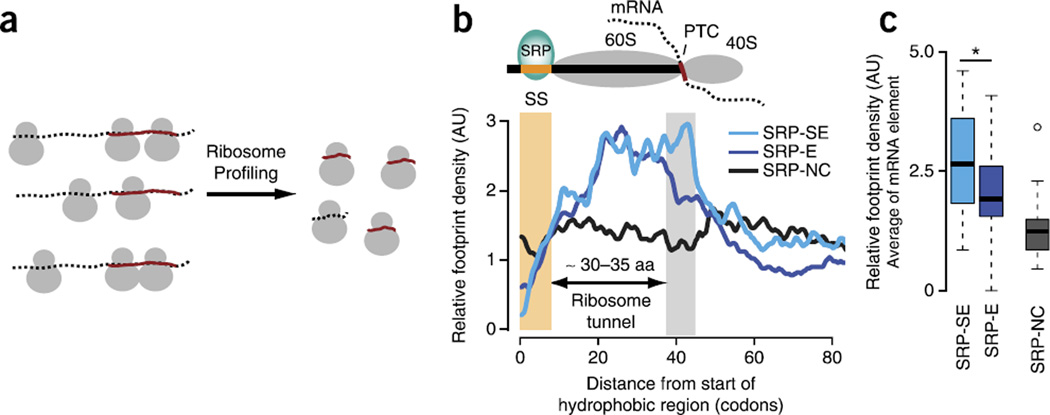
Ribosome profiling confirms a translational slowdown that associates with enhanced SRP binding. (a) Schematic for ribosome profiling. Ribosome profiling experimentally measures local translation kinetics through selective sequencing of ribosome-protected footprints. Higher footprint densities indicate slower local translation kinetics. (b) Median ribosome footprint density profiles (arbitrary units, AU) for SRP-SE (n = 16), SRP-E (n = 33) and SRP-NC (n = 20) substrates. Only SRP-SE substrates exhibit higher footprint densities in the translational-slowdown element (gray bar) downstream of the SS (yellow bar). Aa, amino acids. (c) Distributions of average relative footprint densities in the downstream translational-slowdown element for individual SRP-SE, SRP-E and SRP-NC proteins. SRP-SE substrates are locally translated more slowly compared to SRP-E and SRP-NC substrates just when the hydrophobic SS is exposed outside the ribosome (P = 0.042 by Wilcoxon rank-sum test; n values as in b). Box plots show the data distribution through median (center line), first and third quartiles (filled box), 1.5× interquartile range (dashed line) and extreme values (circles). *P < 0.05.