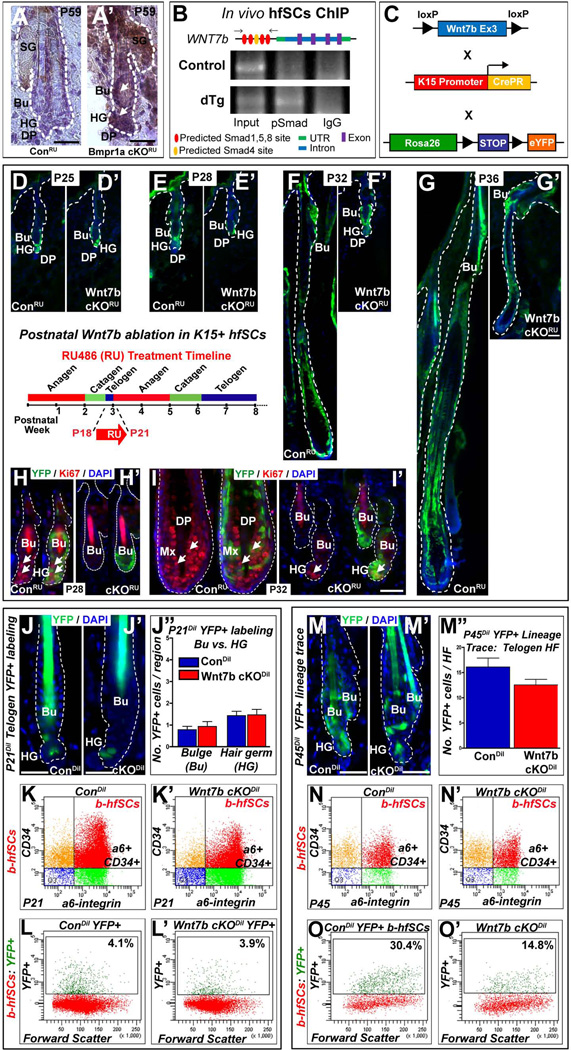
Figure 6. Delayed hair follicle stem cell (hfSCs) activation following Wnt7b ablation.
(A’) Up-regulation of WNT7b in telogen bulge (Bu) hfSCs and hair germ (HG) after BMP pathway inactivation in BMPR1A cKORU at P59 compared to ConRU HFs (A) where WNT7b staining was absent. (B) In vivo hfSCs ChIP PCR reveals selective precipitation of DNA fragments that possess canonical Smad binding elements of Wnt7b promoter, flanking primers (arrows) in schematic (C) Schematic of transgenic mice generation with targeted Wnt7b deletion in the bulge: Con and Wnt7bfl/fl transgenic mice (both K15CrePR+; YFP+) were treated with RU486 from P18–P21 (schematic inset) to YFP label K15+ control (ConRU) and ablate Wnt7b in Wnt7b cKORU hfSCs. (D–G’) The following postnatal hair cycle was monitored. (D, D’) After 4 days (P25), both ConRU and Wnt7b cKORU HFs remained in telogen due to RU treatment 35. (E) By P28, YFP+ ConRU HFs displayed telogen-anagen transition with enlarged, activated HG, however, YFP+ Wnt7b cKORU HFs remained in telogen (E’). At P32, YFP+ ConRU HFs were in full anagen with efficient YFP+ labeling throughout all HF layers (F), however, YFP+ Wnt7b cKORU displayed enlarged HGs indicating initial HF activation and early telogen-anagen transition (F’). By P36, ConRU YFP+ HFs were in full anagen with HFs extending deep into the dermis (G), whereas, Wnt7b cKORU YFP+ HFs were in early anagen (G’). (H–I’) Immunofluorescence for the cell proliferation marker, Ki67 in YFP+ ConRU and Wnt7b cKORU HFs. (H) At P28, ConRU YFP+ HFs display strong nuclear Ki67 staining in the HG and lower Bu cells indicating HF activation, (H’) however, Wnt7b cKORU HFs lacked Ki67 staining. (I) At P32, nuclear Ki67 staining was detected in the matrix cells of YFP+ ConRU HFs. (I’) Wnt7b cKORU YFP+ HFs displayed nuclear Ki67 staining in the HG indicating early HF activation. DAPI (blue) counterstaining was used to label all nuclei. Representative images of P21 ConDil (J) and Wnt7b cKODil (J’) YFP+ labeled HFs and quantification of the number of YFP+ cells per Bu and HG compartment (J”). (K–K’) FACS analysis employing α6-integrin and CD34 staining with α6-integrinHigh/CD34High basal hfSCs population (b-hfSCs, red) in ConDil and Wnt7b cKODil at P21 HFs. FACS analysis of b-hfSCs (α6High/CD34High) gated population for YFP positive cells in Con (L) and Wnt7b cKO (L’) at P21 HFs. Representative images of P45 ConDil (M) and Wnt7b cKODil (M’) YFP+ labeled HFs and quantification of the number of YFP+ cells per HF (M”). (N–N’) FACS analysis of α6-integrin and CD34 positive b-hfSCs population (b-hfSCs, red; α6-integrinHigh/CD34High) in ConDil and Wnt7b cKODil at P45 HFs. FACS analysis of b-hfSCs (α6High/CD34High) gated population for YFP positive cells in ConDil (O) and Wnt7b cKODil (O’) at P45 HFs. Scale bar = 50µm.