Summary
The interaction of the serine/threonine kinase Pelle and adaptor protein Tube through their N-terminal death domains leads to the nuclear translocation of the transcription factor Dorsal and activation of zygotic patterning genes during Drosophila embryogenesis. Crystal structure of the Pelle and Tube death domain heterodimer reveals that the two death domains adopt a six-helix bundle fold and are arranged in an open-ended linear array with plastic interfaces mediating their interactions. The Tube death domain has an insertion between helices 2 and 3, and a C-terminal tail making significant and indispensable contacts in the heterodimer. In vivo assays of Pelle and Tube mutants confirmed that the integrity of the major heterodimer interface is critical to the activity of these molecules.
Introduction
The dorsal–ventral axis of the Drosophila embryo is established by maternal effect genes that direct the spatially graded transfer of Dorsal, a Rel family NF-κB homolog, from the cytoplasm to nuclei at the syncytial blastoderm stage (Belvin et al., 1995). The events that lead to nuclear localization of Dorsal are initiated at the plasma membrane, through recruitment of the serine/threonine kinase Pelle upon activation of the transmembrane receptor Toll. It is believed that Pelle is recruited to the plasma membrane by the adaptor protein Tube (Großhans et al., 1994; Galindo et al., 1995; Towb et al., 1998), which itself translocates to the membrane upon Toll activation. Acting through a kinase cascade, Pelle effects the phosphorylation and consequent degradation of Cactus, an IκB homolog that otherwise sequesters Dorsal in the cytoplasm (Reach et al., 1996). Coupling through different Rel family transcription factors, Pelle and Tube also mediate the innate immune response of Drosophila toward fungal infection (Lemaitre et al., 1996). In mammals, paralogs of these proteins have been identified that mediate a variety of cellular and innate immune responses (Medzhitov and Janeway, 1998; Hoffmann et al., 1999).
The amino-terminal interaction modules of Pelle and Tube show limited amino acid sequence similarity to death domains, a family of folds identified in proteins that assemble components of apoptotic signaling pathways (Tartaglia et al., 1993; Feinstein et al., 1995). These include the TNF receptor and Fas, receptor-associated proteins TRADD and FADD, and effector proteases such as Caspase 8 (Baker and Reddy, 1998). Death domains are generally capable of homotypic interactions. Structural studies of the death domains of Fas (Huang et al., 1996), P75 neurotrophin receptor (Liepinsh et al., 1997), and FADD (Jeong et al., 1999) reveal a common helical fold that is also characteristic of death effector domains (DED) (Eberstadt et al., 1998) and caspase recruitment (CARD) domains (Chou et al., 1998; Qin et al., 1999). However, death domains display considerable sequence diversity, consistent with the various cellular functions in which they participate. The apparent absence of a conserved interaction surface suggests that death domains may associate by a variety of mechanisms.
Here, we describe the crystal structure of a death domain heterodimer formed by the death domains of Pelle and Tube. We have evaluated the functional relevance of the dimer interface by an in vivo assay, using site-directed mutants of Pelle and Tube to rescue Drosophila embryos carrying null mutations in pelle or tube (Letsou et al., 1991; Shelton and Wasserman, 1993). We describe a mode of interaction that may be unique to Tube and Pelle and yet is supported by a scaffold common to all death domains.
Results and Discussion
Complex Formation, Crystallization, and Structure Determination
Amino-terminally hexahistidine-tagged death domains of Pelle (residues 26–129, Pelle-DD) and Tube (residues 1–184, Tube-DD) from D. melanogaster were expressed as recombinant proteins in E. coli. Histidine tags were removed from both proteins before the final purification step. A stoichiometric complex of Pelle-DD:Tube-DD can be obtained by coincubating the two proteins. The apparent molecular mass of the complex, as determined by size exclusion chromatography and equilibrium ultra-centrifugation, is consistent with a Pelle-DD:Tube-DD dimer (data not shown). The dimer is stable and does not dissociate upon subsequent rechromatography.
Crystals of Pelle-DD:Tube-DD grow as rectangular prisms in space group P212121 and contain two molecules of each domain in the asymmetric unit (Table 1). Diffraction from these crystals can be detected to 1.9 Å resolution using a high-intensity synchrotron source. The structure was determined by the multiwavelength anomalous dispersion (MAD) method (Hendrickson, 1991), using isomorphous crystals produced with selenomethionyl Pelle-DD and Tube-DD. Crystallographic phases were determined using a four-wavelength MAD data set measured at the Advanced Light Source (Berkeley, CA) from crystals maintained at 100 K (Table 1). The resulting electron density map was of excellent quality, and all four protein monomers in the asymmetric unit could be traced without ambiguity. The model has been refined with data extending to 2.0 Å to working and free R factors (Brünger, 1992) of 0.21 and 0.24, respectively (Table 1). The final atomic model consists of two molecules of Pelle-DD (P1, residues 28–129; P2, residues 26–129; in addition to four N-terminal residues, GSHM, derived from the expression vector), two molecules of Tube-DD (T1, residues 23–172; T2, residues 23–175), an ordered HEPES molecule, and 266 water molecules.
Table 1.
Data Collection, Phasing, and Refinement Statistics
| Space group | P212121 | |||||
|---|---|---|---|---|---|---|
| Unit cell | a = 58.123 Å, b = 87.495 Å, c = 117.662 Å | |||||
|
| ||||||
| Dmin (Å ) | Unique Reflections |
Redundancy | Completeness (%)a | <I>/<σ1< | Rsym (%)b | |
| SeMet λ (0.9808 Å) | 2.1 | 69,355 | 3.7 | 99.7 (98.9) | 24.8 (3.4) | 4.4 (37.5) |
| SeMet λ2 (0.9801 Å) | 2.1 | 69,558 | 3.7 | 99.7 (98.9) | 24.2 (3.2) | 4.8 (39.2) |
| SeMet λ3 (0.9799 Å) | 2.1 | 68,951 | 3.7 | 99.7 (99.1) | 24.5 (3.8) | 4.7 (34.0) |
| SeMet λ4 (0.9500 Å) | 2.1 | 67,894 | 3.7 | 99.8 (99.3) | 22.5 (2.8) | 5.3 (48.2) |
| Native (0.9810 Å) | 2.0 | 41,269 | 3.7 | 98.9 (98.6) | 17.7 (2.4) | 5.5 (49.9) |
| MAD Difference Ratios (25.0-2.1 Å)c | Anomalous Scattering Factors | |||||
|---|---|---|---|---|---|---|
| λ 1 | λ 2 | λ 3 | λ 4 | f′ (e−) | f″ (e−) | |
|
|
|
|||||
| λ 1 | 0.036 | 0.036 | 0.047 | 0.046 | −7.3 | 0.6 |
| λ 2 | 0.064 | 0.038 | 0.056 | −10.9 | 4.1 | |
| λ 3 | 0.078 | 0.048 | −8.9 | 6.2 | ||
| λ 4 | 0.061 | −2.8 | 3.6 | |||
| Phasing powerd and figure of merit (FOM)e | ||||||||
|---|---|---|---|---|---|---|---|---|
| λ1 → λ1− | λ1 → λ2+ | λ1 → λ2− | λ1 → λ3+ | λ1 → λ3− | λ1 → λ4+ | λ1 → λ4− | FOM | |
| 25–2.1 Å | 0.57 | 0.58 | 1.22 | 0.77 | 1.54 | 1.32 | 1.73 | 0.57 |
| 2.2–2.1 Å | 0.23 | 0.04 | 0.45 | 0.24 | 0.62 | 0.27 | 0.40 | 0.23 |
| Refinement | |
|---|---|
| Resolution range for refinement | 25.0–2.0 Å |
| No. of protein atoms | 4388 |
| No. of water atoms | 266 |
| No. of heteroatoms | 15 |
| Rmsd bond lengths | 0.006 Å |
| Rmsd bond angles | 1.067° |
| Rmsd bonded main chain B factors | 1.73 Å2 |
| Rmsd bonded side chain B factors | 2.41 Å2 |
| Rworkf | 21.1% |
| Rfreeg | 24.4% |
| Average B factor | 43.3 Å2 |
Numbers in parentheses correspond to the last resolution shell.
Rsym = ΣhΣi|Ii(h) − <I(h)> |/ΣhΣIi(h), where Ii(h) and <I(h)> are the ith and mean measurement of the intensity of reflection h.
MAD difference ratios are <(Δ|F|)2>1/2/<(|F|)2>1/2, where Δ|F| is the dispersive (off-diagonal elements), or Bijvoet difference (diagonal elements).
MAD phasing power is defined as {<|Fλ1 − Fλi|2>∫φ Pλ1→λi (φ) [||Fλ1| exp (iφ) + Fλi − Fλ1| − |Fλi|]2 dφ}1/2, where Fλ1 is the structure factors at the reference wavelength λ1, Fλi are the structure factors at wavelength λi (indicated by a superscript “+”) or its Friedel mate (indicated by a superscript “−”), φ is the phase angle and Pλ1→λi (φ) is the experimental phase probability distribution.
Figure of merit is defined as <{∫φP (φ) exp (iφ) dφ}/ ∫φ P (φ) dφ>.
Rwork = Σh ||Fobs (h)| − |Fcalc (h) || /Σh |Fobs (h)|, where Fobs (h) and Fcalc (h) are the observed and calculated structure factors, respectively.
Rfree is the R value obtained for a test set of reflections consisting of a randomly selected 10% subset of the complete data set excluded from refinement
Structures of Pelle-DD and Tube-DD
As predicted from their amino acid sequences (Feinstein et al., 1995) (Figure 1), Pelle-DD and Tube-DD possess three-dimensional folds that identify them as death domains (Figure 2A). The same topology characterizes a structural superfamily that includes DED (Eberstadt et al., 1998) and CARD (Chou et al., 1998; Qin et al., 1999). Members of this superfamily possess helical bundle folds with pronounced variation in the length and orientation of helices. Pelle-DD and Tube-DD can be described as antiparallel six-helix bundles or as sandwiches with Greek key connectivity. One face of the sandwich is formed by helices 1, 4, and 6, and the opposite face by helices 2, 3, and 5 (Figure 2A). The cores of the molecules are formed by the helical hairpins α1-α2 and α4-α5, which are wedged into each other like a pair of interlocking scissors. Four of the five most highly conserved residues in the death domain fold, represented by residues L87, W90, L101, and F105 in Pelle (Figure 1), form the hydrophobic nucleus about which the helical hairpins are packed and fix the spatial relationship between them. Indeed, these four residues can be used to obtain a close superposition of available death domain structures (Figure 2B). This helical core is largely preserved in all members of the death domain family, but not in the superfamily that includes DED and CARD. Pelle-DD and FADD-DED are structurally similar within a core limited to α1, α2, α5, and α6 (rmsd of 1.15 Å for 38 Cα pairs). The best superposition between Pelle-DD and RAIDD-CARD is obtained using only α1, α5, and α6 (rmsd of 1.71 Å for 29 Cα atom pairs).
Figure 1.
Sequence Alignment of Death Domains
Listed are death domains of D. melanogaster Pelle (sp|Q05652), C. elegans Pelle (gi|2814202), human IRAK (sp|P51617), IRAK-2 (gi|4504719), human D. melanogaster Tube (sp|P22812), D. virilis Tube (sp|Q08171), Bombyx mori Tube cDNA clone e40070 (gi|4156307), human MyD88 (gi|176309), rat P75 neurotrophin receptor (sp|P07174), human Fas (sp|P25445), mouse FADD (sp|Q61160), human TNF receptor 1 (sp|P19438), and human TRADD (sp|Q15628). Red cylinders indicate D. melanogaster Pelle death domain helices; blue cylinders indicate the two Tube death domain helix inserts. The helices of D. melanogaster Tube death domain, rat P75 receptor, human Fas, and mouse FADD are underlined. Highly conserved residues are highlighted in yellow, and conserved residues are highlighted in green. Residues are colored red, orange, or green where mutations cause complete loss, partial loss, or no obvious loss of function, respectively. The D. melanogaster Pelle or Tube residues at both P:T interfaces (see text) are indicated by red dots below the sequence; those at only one of the interfaces are indicated with orange dots.
Figure 2.
The Six-Helix Bundle Fold of Pelle-DD and Tube-DD
(A) P2:T2 dimer is shown with P2 in magenta and T2 in green. The inserted helices αA and αB of Tube-DD are colored cyan.
(B) Superposition of Pelle-DD (magenta) and Fas-DD (gray). The view for Pelle-DD is the same as in (A). The five highly conserved residues are displayed as ball-and-stick models with those of Pelle-DD indicated by arrows. Only four residues, L87, W90, L101, and F105, are used to obtain this superposition. The helices of Fas-DD are marked.
(C) P2:T2 dimer is superimposed onto P1:T1 using the Pelle-DD molecules of the respective dimers. P1, T1, P2, T2, and T2A are colored red, cyan, magenta, and green, respectively. The helices of T1 molecules are labeled.
Figures prepared using Gl_render (Esser, 1999), BOBSCRIPT (Esnouf, 1997), MOLSCRIPT (Kraulis, 1991), and POV-ray (1998).
As observed in the solution structures of Fas, FADD, and P75, α3 of Pelle-DD and its counterpart in Tube-DD appear to be flexible with high thermal parameters. The position of this helix, which projects from the core of the protein, is determined in part by the highly conserved residue W55 (of Pelle-DD) in α2. Pelle W55 acts as a wedge between α3 and the rest of the structure. Fas is a notable exception, where W55 is replaced by the smaller hydrophobic residue valine and α3 is rotated into the core of the domain relative to the corresponding helices in Pelle-DD, Tube-DD, and FADD-DD (Figure 2B). The distance of α3 to the rest of the death domain dictates the curvature and convexity of the local molecular surface and may have structural implications for molecular recognition.
Tube-DD contains elements not present in Pelle-DD or in other members of the death domain family. Inserted between α2 and α3 is a compact Ω loop (Leszczynski and Rose, 1986) containing two short helices (αA and αB), which packs against α4 at the narrow end of the bundle. As discussed below, this insert blocks a potential intermolecular interaction surface. The amino acid sequence corresponding to this insert is poorly conserved among the D. melanogaster, D. virilis, and B. mori orthologs of Tube (Figure 1). Tube-DD also possesses a C-terminal extension that is not present in other death domains but that may have an analog in the MyD88 death domain, a vertebrate functional homolog of Tube-DD (Muzio et al., 1997; Medzhitov et al., 1998). A note-worthy feature of this segment is a right-handed loop, beginning at residue 161 and composed of the sequence PVDGPGA. Both proline residues are trans, and G164 adopts a main chain conformation (Φ = 95° and 93°, and Ψ = 178° and 177°, for T1 and T2, respectively) that is accessible only to glycine. The GPG sequence is rigid (average B factor for these residues is 36 Å2, whereas the average B factor for all protein atoms is 43 Å2) and contributes to a critical Pelle-DD:Tube-DD interface, as described below.
The electrostatic potential of Pelle-DD and Tube-DD, computed at their respective solvent-accessible surfaces, reveals discrete and interspersed patches of high electrostatic potential in both domains (Figure 5A). A groove formed by the α4-α5 and α2-α3 hairpins of Pelle-DD is positively charged.
Figure 5.
The C-Terminal Tail of Tube-DD Contributes Significant Interaction Surface
(A) The P2:T2 dimer: P2 is displayed as electrostatic surface (left) and magenta ribbon (right), T2 as green ribbon (left) and electrostatic surface (right), respectively. The color scale for the charge distribution extends from −10 kT/e− (red) to +10 kT/e− (blue) calculated using GRASP (Nicholls et al., 1991). The T2 carboxy-terminal hydrophobic residues in contact with P2, as well as charged residues of P2 surrounding the above region, are labeled on the respective surfaces. The T2 C-terminal tail is shown as a ball-and-stick model starting from residue 161 on the left.
(B) The various contacts at the dimer interface are displayed as a schematic diagram with Pelle residues shaded magenta and Tube, green. Hydrogen bonds, salt bridges, and van der Waals interactions are indicated by black arrows (donor to acceptor), red arrows, and dotted lines, respectively.
The Pelle-DD:Tube-DD Dimer Interface
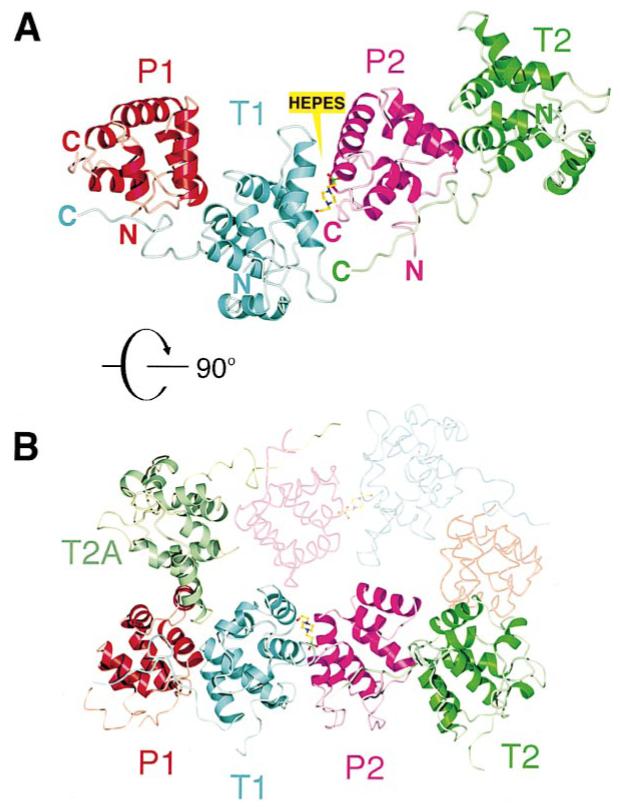
There are four molecules in the crystallographic asymmetric unit that we designate as P1, P2, T1, and T2. A Tube molecule from another asymmetric unit inserts its α3 helix and the following loop between P1 and T1 and is distinguished as T2A molecule (Figure 3B). The four molecules of the asymmetric unit are arranged in the sequence P1:T1:P2:T2 and form an approximately linear array. Since the crystal was formed from a solution of Pelle-DD:Tube-DD dimers, we conclude that the P:T interface is the basis of the Pelle-DD:Tube-DD complex formed in vitro. Structural, biochemical, and genetic evidence discussed below all support this view.
Figure 3.
Interfaces between Pelle-DD and Tube-DD
(A) The four death domains of the crystallographic asymmetric unit are colored according to Figure 2C.
(B) The asymmetric unit is rotated 90° about the horizontal axis with an additional Tube (T2A) from another asymmetric unit (shown in low contrast) colored pale green.
The two P:T interfaces, as measured by occluded area of solvent-accessible surface and the number of noncovalent interactions, are the most extensive of the intermolecular contacts observed in the lattice (Table 2). The contact surface is roughly orthogonal to the axis of the helical bundles (Figure 2A). Insertion of T2A into the P1:T1 interface obviates certain P:T contacts that are present in the P2:T2 interface (Figures 3B and 4). In addition, the C-terminal residues of T2 adopt a different conformation than those in T1, allowing a more intimate contact with Pelle-DD in P2:T2 than in P1:T1. Consequently, although the two dimers are similar, P2:T2 forms a more extensive interface (Table 2). Subsequent description of the P:T interface focuses on this dimer.
Table 2. Drosophila Embryogenesis Phenotypes Generated by RNA Injection Assay
| A. Mutants for Pelle | ||
|---|---|---|
|
| ||
| RNA Injected | Pelle Activity | Protein Expressed |
| Uninjected | − | − |
| Δ2-25 | +++ | ND |
| S73Y | +++ | ND |
| Wild type | +++ | + |
| P:T (P1:T1, 1700 Å2; P21:T2, 2180 Å2) | ||
| R35E | − | + |
| Tube E50K | − | −/+ |
| R35E; Tube E50K | + | ND |
| H96A | −/+ | + |
| K107D | + | + |
| D67R | ++ | ND |
| Q93G | ++ | ND |
| Q93W | +++ | ND |
| H96E | ++ | ND |
| D67K | +++ | ND |
| G92W | +++ | ND |
| H96R | +++ | ND |
| T2A:(P1:T1) (1050 Å2) | ||
| D67R | ++ | ND |
| D67K | +++ | ND |
| T1:P2 (910 Å2) | ||
| D50K | − | − |
| R115D | +++ | ND |
| B. Mutants for Tube | ||
|---|---|---|
|
| ||
| RNA Injected | Tube Activity | Protein Expressed |
| Uninjected | − | − |
| Δ62–89 | − | ND |
| Δ4–22 | + | ND |
| Δ68–71 | +++ | ND |
| D97R, E98R | +++ | ND |
| Wild type | +++ | + |
| P:T (P1:T1, 1700 Å2; P21:T2, 2180 Å2) | ||
| E50K | − | −/+ |
| E140K | − | + |
| S143I, A144T | − | + |
| G164E | −/+ | + |
| N51W | + | ND |
| T2A:(P1:T1) (1050 Å2) | ||
| D106K | −/+ | + |
| T1:P2 (910 Å2) | ||
| D38I | + | ND |
| D115R | ++ | ND |
The list ofmutants ([A] for Pelle and [B] for Tube) is sorted according to interface (see text). The buried solvent-accessible surface area at each interface is indicated. Mutants are named as follows: starting amino acid, position, and amino acid after mutagenesis or, for deletions (Δ), by the first and last residue removed. As indicated, some constructs contain two mutations. Activity is defined as follows: (−) no rescue, no dorsoventral (D/V) patterning elements formed; (−/+) 10%–50% of embryos exhibit dorsal–lateral patterning elements (filzkörper), no hatching; (+) hatch rate 1%–5%, greater than 90% of embryo cuticles show D/V patterning defects; (++) hatch rate 6%–20%, greater than 60% of embryo cuticles show D/V patterning defects; (+++) hatch rate 20%–70%, 20%–60% of cuticles show D/V patterning defects. Protein expression is quantified as follows: (−) no expression detected by immunoblot analysis; (−/+) protein expression is 30%–50% of wild-type levels; (+) protein expression is identical to the expression from a wild-type construct; ND, not determined.
Figure 4.
Detailed View of the Dimer Interfaces
(A) The P2:T2 dimer interface with interacting residues is displayed as a ball-and-stick model. Hydrogen bonds and salt bridges are indicated by gray dots. For clarity, part of the T2 C-terminal tail is omitted. The molecules are colored as in Figures 2C and 3.
(B) The corresponding interface between P1 and T1 is shown with the intercalated T2A molecule. Part of the T1 C-terminal region is omitted for clarity.
Pelle-DD and Tube-DD contact each other at two distinct but adjacent sites in P:T dimers. The first of these is created by the insertion of α4 and the following loop of Pelle into a groove of Tube. One side of the groove is formed by the α1-α2 corner, the other by α6 and preceding loop (Figure 4A). Within this cradle, the C terminus of Pelle α4 directly abuts the N terminus of Tube α6. These two elements are connected by a pseudo-helical hydrogen bond between the main chain amide of Tube S143 and the carbonyl oxygen of Pelle Q93. The side chain hydroxyl of Tube S143 also forms a hydrogen bond with the carbonyl oxygen of Pelle G92, and that of Tube D146 interacts with the side chain of Pelle N95. There are, in addition, several polar or charge–charge interactions between residues in the α1-α2 corner of Tube and residues in Pelle α1 (Tube E50–Pelle R35) and α3 (Tube N51–Pelle Q71; Tube K56–Pelle D67) (Figures 4A and 5B).
The second interaction site centers on the insertion of the GPG sequence at the C-terminal tail of Tube-DD into a cavity between the α4-α5 and α2-α3 hairpins of Pelle. Proline 165 of Tube forms numerous van der Waals interactions with Y94 of Pelle α4 and a main chain hydrogen bond to Pelle W90 and H96. Pelle Y94 is also hydrogen bonded to Tube E140 and Pelle Q93, thus essentially connecting the two interaction sites into one continuous interface. On the opposite side of the same groove, Tube P165 makes van der Waals contacts with V62 and L64 of Pelle α2. The C-terminal extension of Tube inserts three hydrophobic residues (I169, L171, and L173) into a cavity between α5 and the N terminus of the Pelle-DD and participates in several main chain–side chain hydrogen bonds (Figure 5). Negatively charged residues at the C terminus of the Tube extension are disordered in the structure but would be positioned near positive charges in the Pelle α5-α6 cleft. In total, there are 14 direct hydrogen bonds and more than 100 van der Waals interactions at the P2:T2 interface.
Comparison of the Two Dimers in the Crystallographic Asymmetric Unit
Superposition of P1:T1 onto P2:T2 (Figure 2C) demonstrates that the P:T interface is plastic. Structural differences between the two are due to the insertion of a second Tube molecule (T2A) into the P1:T1 interface. T1 accommodates this intrusion by executing a rigid body rotation about a fulcrum located near P165 in the GPG loop. This rotation allows P105 of T2A to intercalate between I60 and K56 of T1 (Figure 4B). With respect to Pelle, the orientations of Tubes in the two P:T dimers differ by a 7° rotation about the P→T axis passing through in the two Tube P165 (Figure 2C). Preserved P:T dimers are the contacts surrounding the GPG loop of Tube and the direct hydrogen bonds between Pelle α4 and Tube α6. In contrast, some polar residues further from the rotation axis find alternative interaction partners. The ion pair between Tube K56 and Pelle D67 in P2:T2 is replaced in P1:T1 by a salt bridge with D106 of T2A. The hydrogen bond between Pelle Y94 and Tube E140 and that between Pelle N95 and Tube D146 are lost in P1:T1 (Figure 4). Isoleucine 169 and L171 of Tube also occupy different hydrophobic environments in the two dimers, in part because the C-terminal residues of Tube (following P165) adopt alternative conformations.
In the crystal lattice, the P1:T1 and P2:T2 dimers are related by translation and a rotation that tilts the P→T axes of the two dimers 60° with respect to each other (Figure 3A). A strictly translational relationship between T1 and P2 is blocked by the extended loop formed by residues 120–128 of T1. The T1:P2 contact area contains a bound molecule of HEPES buffer that accounts for a substantial portion of the interaction surface. This, together with the paucity of direct and specific contacts between the domains and absence of Pelle-DD:Tube-DD aggregate formation in solution, indicates that the T1:P2 interaction is not stable.
Molecular Genetic Analysis of the Domain Interfaces
To evaluate the relevance of lattice interfaces to the function of Pelle and Tube in vivo, we have introduced point mutations designed to destabilize or abolish interactions at selected interfaces. These mutations affect sites on the surface of monomeric domains, minimizing the likelihood of their altering protein structures. The ability of mutant Pelle or Tube molecules to rescue Drosophila embryos, in which the corresponding gene is functionally deleted, can be determined by injecting RNA encoding the mutant molecules and scoring specific markers for dorsal–ventral axis formation (Anderson and Nüsslein-Volhard, 1984) (Table 2). In such assays, the strength of the phenotype is a qualitative measurement of the signaling activity of the mutant Pelle or Tube molecules. If Pelle or Tube is absent or if they are unable to interact with one another, only the most dorsal patterning elements are formed. A weak Pelle:Tube interaction that supports low levels of signaling will result in the formation of dorsal–lateral patterning elements, such as filzkörper. Higher levels of Pelle:Tube activity are required to develop ventral–lateral patterning elements, including ventral denticles.
Phenotypes generated by RNA injection of mutant Pelle and Tube show that residues at the P:T interface are required for activity in vivo (Table 2). The C terminus of Tube-DD, which forms the rigid fulcrum, appears to be essential. A Tube mutant in which all residues following N154 are deleted fails to restore embryonic development in tube mutant eggs (Letsou et al., 1993). Likewise, truncation of the Tube-DD at D163 (preceding GPG) abolishes its ability to form stable dimers with Pelle under the conditions used to prepare complexes for crystallization (data not shown). An EMS-induced mutation in tube that results in a fully dorsalized phenotype converts E140 to lysine (Letsou et al., 1993). Located at the P:T interface, E140 forms hydrogen bonds to Pelle Y65 and Y94 and may also stabilize the GPG turn. Mutation of Tube G164 to glutamate, which is expected to disrupt the conformation of the GPG turn, likewise renders Tube inactive. Mutation of Tube S143, at the core of the interface, is fully inactivating, and substitution of Pelle H96, the hydrogen bonding partner of Tube P165, by alanine is debilitating. Charge-reversal mutations of either Tube E50 or Pelle R35, which form an ion pair, also abolish activity. All of the mutant proteins described above are expressed in Drosophila embryos (Table 2), consistent with the hypothesis that these mutations disrupt protein–protein interactions but not protein folding and structure.
By coinjecting mutant Tube and Pelle RNAs, we next assayed whether a mutation at the surface of the interface contributed by one death domain could be compensated by a reciprocal alteration in the pairing partner. For this analysis, we chose the mutations converting Pelle R35 to glutamate and Tube E50 to lysine. Although each mutation is inactivating individually, the two mutations together are predicted to restore an ion pair. Indeed, coinjection of the two mutants restores significant levels of signaling to pelle mutant embryos, as evidenced by strong rescue (ventral denticles and muscle movement). These experiments provide direct confirmation that Pelle and Tube associate in vivo through the P:T interface.
The plasticity of the P:T interface, apparent in the differences between P2:T2 and P1:T1, offers an explanation for the fact that some mutations at the P:T interface do not abolish Pelle or Tube function. For example, substitution of Pelle Q93G or G92W, which might be expected to sterically disrupt the P:T contact, leads to only weak or wild-type phenotypes. All of the benign mutations at the P:T interface (Table 2) could be accommodated by a rotation of Tube-DD (with respect to Pelle-DD) similar to that which transforms the P2:T2 dimer to P1:T1 (Figure 2C). In contrast, mutations at sites of the interface unaffected by this apparent rotation (i.e., near the axis of rotation) are inactivating.
Death Domain Interactions in Dorsal and NF-κB Signaling
The P:T dimer interface incorporates both rigid and plastic elements. Surprisingly, the most rigid contacts appear to involve Tube residues that lie C-terminal to its death domain interacting with the surface of Pelle-DD. A more plastic interface results from direct interactions between helical elements of the two domains, centered about an end-to-end contact between the α4 helix of Pelle-DD with α6 of Tube-DD. As a potential phosphorylation site by Pelle (Z. Liu, unpublished data), residue S143 of Tube α6 is at the center of the dimer interface. This is consistent with the observation that inactivation of Pelle kinase activity leads to more persistent Pelle: Tube interactions (Edwards et al., 1997). The dimer interface does not obstruct residues C-terminal to the death domains of either Pelle or Tube, suggesting that the kinase domain of Pelle and the C-terminal domain of Tube are unhindered by the formation of the complex. In the crystal lattice, the P:T interface can be partially disrupted by insertion of a second Tube-DD T2A. Although the interface between the second Tube-DD and the P:T dimer is small, it may be of some functional significance, if, for example, Tube aggregation accompanies Toll signaling (Towb et al., 1998; Großhans et al., 1999). Tube-DD self-association could not resemble the interaction seen in the P:T interface, because the Tube insertion between α2 and α3 occupies the binding site for the C terminus of a second Tube-DD.
The P:T interface may be recapitulated in homologs of Pelle and Tube. In vertebrates, activation of NF-κB by either interleukin receptor 1 (IL-R1) (Muzio et al., 1997) or the human Toll receptors (Medzhitov et al., 1998) proceeds through formation of a complex between the death domain of IRAK, a Pelle ortholog, and MyD88, a Tube counterpart. Like Tube, MyD88 is bipartite with an amino-terminal death domain (Figure 1). Critical to complex formation with the IRAK death domain is an intermediate domain of MyD88 located between its C-terminal TIR and death domains (Medzhitov et al., 1998). The MyD88 intermediate domain is analogous to the residues C-terminal to the Tube-DD, which form an essential contact with Pelle. However, the intermediate domain of MyD88 contains a series of charged and polar residues in place of the GPG sequence of Tube-DD and is unlikely to fold in a conformation similar to that in Tube. Further, whereas MyD88 binds directly to the cytoplasmic TIR domain of hToll, direct interaction between Tube and Drosophila Toll has not been demonstrated (Galindo et al., 1995; Edwards et al., 1997).
Diverse Association Mechanisms in the Death Domain Superfamily
The P:T dimer has little in common with the dimeric arrangements proposed or demonstrated for death, death effector, or CARD domains. First, the α2-α3 surface, implicated in homo- and heterodimeric complexes of the death domains of Fas (Huang et al., 1996), FADD (Jeong et al., 1999), and the CARD of apoptotic protease-activating factor 1 (Apaf1) (Qin et al., 1999), plays no major role in the formation of the P:T dimer, which instead relies on contacts between α4-α5 (Pelle-DD) with α6 (Tube-DD). Further, although mutations of residues in α3 of Fas compromise function, they have little effect upon the biological activity of Pelle and Tube (Figure 1 and Table 2). Of note, however, the N terminus of α2 (H2) in Apaf1 directly abuts the C terminus of H1b in procaspase 9 CARD, forming a helix–helix contact roughly similar to that between the α4 (Pelle-DD)–α6 (Tube-DD) interaction. Death domains (in distinction to DED and CARD domains) participate in multiple homomeric and heteromeric interactions and hence may employ several binding surfaces, as mutagenic studies of TRADD (Park and Baichwal, 1996) and TNFR1 (Tartaglia et al., 1993) appear to suggest.
Whereas Fas-DD:FADD-DD heterodimers are proposed to be isologous (i.e., involving corresponding structural elements in mutual contact; Jeong et al., 1999), the association between Pelle-DD and Tube-DD is an open-ended, “head-to-tail” interaction, involving opposite or adjacent surfaces with reference to the common fold of the two monomers. In this respect, the Pelle-DD:Tube-DD interaction is similar to that proposed for the interaction of procaspase 2 with RAIDD and demonstrated in crystals of the Apaf1-CARD:procaspase 9–CARD complexes (Chou et al., 1998; Qin et al., 1999).
It is now apparent that the interaction surfaces for death domains vary substantially. The proposed isologous interactions between Fas-DD and FADD-DD, as well as the heterologous contacts between CARD domains, involve contact between oppositely charged surfaces. The DED of FADD, in contrast, is proposed to form contacts with the complementary DED of FLICE/Caspase 8 at a predominantly hydrophobic interface (Eberstadt et al., 1998). Lastly, the death domains of Pelle and Tube exhibit no well-defined charged patches, and the interactions between them involve a mixture of polar, charged, and hydrophobic interactions. In addition, the major interface between Pelle-DD and Tube-DD incorporates a critical surface that is extraneous to the death domain itself. Indeed, Tube-DD, if stripped of the residues C-terminal to the α6 of the death domain proper, both fails to form complexes with Pelle-DD in vitro and is biologically inactive. The importance of death domain bordering residues has been demonstrated for MyD88 protein, as mentioned above, and is underscored by the observation that deletion of nine residues N-terminal to the death domain of Fas abrogates its ability to bind FADD (Chang et al., 1999).
The crystal structure of Pelle and Tube death domains in association reveals a predominant mode of dimerization, sufficient to rationalize the mechanism through which Pelle is sequestered to its site of action by Tube. At the same time, other nonexclusive modes of Pelle-DD:Tube-DD (and Tube-DD:Tube-DD) interactions are observed. These interactions may demonstrate mechanisms by which new interaction surfaces are formed by death domain dimerization. Recruitment of TRADD by TNF receptor oligomers (Boldin et al., 1995), for example, would require such a mechanism. The variety of death domain contacts exhibited by both Pelle and Tube, together with observed and deduced interactions of other death domains, fails to identify a single, canonical mode of death domain interaction. Rather, a scaffold common to the death domain superfamily appears to have diverged to allow the assembly of a variety of signaling complexes.
Experimental Procedures
Cloning and Expression
Molecular cloning procedure was carried out according to Sambrook and colleagues (Sambrook et al., 1989) unless stated otherwise. Mass spectroscopic analysis and dideoxynucleotide sequencing were performed by the HHMI core facilities at the University of Texas Southwestern Medical Center. cDNA constructs encoding D. melanogaster Pelle-DD (residues 1–129) and Tube-DD (residues 1–184) were obtained from Dr. Zhiping Liu. An N-terminal truncation mutant of Pelle-DD encoding residues 26–129 (ΔN25Pelle-DD, here-after referred to as Pelle-DD) was created by polymerase chain reaction (PCR) and subcloned into vector pET15b (Novagen) and transformed into BL21 (DE3) cells. The sequences of the PCR primers are available upon request. The above construct and the Tube-DD construct mentioned below both have thrombin cleavable N-terminal hexahistidine tags to facilitate purification and subsequent removal of the tag.
Transformed cells were grown in LB medium in the presence of 100 mg/l ampicillin. Cells were grown at 37°C in LB medium, then induced with 0.5 mM isopropyl-1-thiogalactopyranoside (IPTG) at an optical density (A600nm) of ~1.0. Induction was continued at room temperature for 5 hr. The cells were harvested by centrifugation, frozen in liquid nitrogen, and stored at −80°C.
The Tube-DD (residues 1–184) coding sequence in vector pGEX-4T1 was amplified by PCR and subcloned into pET28a vector (Novagen) and transformed into HMS174 (DE3) (Novagen) cells. The large-scale expression protocol used to express the pET28a-Tube-DD (residues 1–184) in HMS174 (DE3) cells was essentially the same as that of Pelle-DD described above.
Purification and Complex Formation
All purification steps were carried out on ice or at 4°C. Cells were lysed and purification of hexa-His tagged protein by Ni2+-nitrilo-triacetic acid (Ni-NTA) resin was carried out according to the QIA-expressionist protocol (Qiagen). Frozen BL21 (DE3) cells expressing Pelle-DD were thawed in lysis buffer (50 mM Tris·Cl [pH 8.5], 300 mM NaCl) and lysed by sonication (Fisher Scientific Sonic Dismembrator 550). The lysate was centrifuged for 1 hr at 100,000 g, and the supernatant was loaded onto a Ni-NTA column preequilibrated with the lysis buffer. The column was washed with lysis buffer followed by the lysis buffer plus 20 mM imidazole. The protein was eluted with lysis buffer plus 500 mM imidazole (pH 8.5), then dialyzed against thrombin digestion buffer (50 mM Tris·Cl [pH 8.5], 150 mM NaCl, 2.5 mM CaCl2). Thrombin (Boehringer Mannheim) digestion at a ratio of 1:20 (thrombin:Pelle-DD, w/w) at 4°C proceeded for 5 hr and was stopped with 2.5 mM ethelenediamine-tetraacetic acid (EDTA). After dialysis against loading buffer (20 mM Tris·Cl [pH 7.5], 1 mM DTT, and 1 mM EDTA), thrombin-treated protein was subjected to MonoS cation exchange chromatography (Amersham Pharmacia Biotech.). Pelle-DD was eluted with a linear 0 mM to 1 M NaCl gradient. Pelle-DD was concentrated in elution buffer to 10 mg/ml using a Centriprep 3 concentrator (Amicon). Final yield of Pelle-DD is 10 mg/5 g wet cells. The protein was flash frozen with liquid nitrogen in 50 μl aliquots and stored at −80°C.
Tube-DD was purified as above with the following modifications. Thrombin digestion was carried out at a 1:5 (w/w) ratio of thrombin:Tube-DD at 4°C for 5 hr. After digestion, protein was loaded onto a MonoQ anion exchange column preequilibrated with elution buffer, and Tube-DD was eluted at 150 mM NaCl. Fractions containing Tube-DD were subjected to gel filtration chromatography in loading buffer containing 100 mM NaCl to eliminate aggregated protein. Final yield of Tube-DD is about 3–4 mg/5 g wet cells. Recombinant Pelle-DD and Tube-DD contain four and three residues derived from the expression vector, GSH(M), at their N termini, respectively.
To form the Pelle-DD and Tube-DD complex, a 1.1:1 molar ratio of the above purified Pelle-DD and Tube-DD was pooled and dialyzed against the elution buffer (100 mM NaCl), then loaded onto two tandemly connected Superdex-75 and Superdex-200 size exclusion columns (Amersham Pharmacia Biotech.). The fractions containing both Pelle-DD and Tube-DD were eluted with an apparent molecular weight of 29 kDa, which corresponded to a Pelle-DD:Tube-DD dimer. Pooled fractions were concentrated with a Centriprep 10 device in the above elution buffer to 14 mg/ml, frozen, and stored as described above.
Se-Met proteins were produced as above with the following modifications. Methionine auxotrophic B834 (DE3) cells (Novagen) were used for the expression of both Pelle-DD and Tube-DD. The cells were grown in selenomethionine-containing minimal media (Sambrook et al., 1989) supplemented with the other 19 amino acids (Yang et al., 1990). DTT (2–5 mM) was present at each purification step except the Ni-NTA column and thrombin digestion steps. The Se-Met proteins behaved similarly to the native proteins during the purification steps, and the yields of the proteins were similar. The Se-Met protein of Pelle-DD and Tube-DD complex was concentrated in elution buffer with 100 mM NaCl and 5 mM DTT with a Centriprep 10 to 10 mg/ml and stored at −80°C as described above. Se-Met proteins are somewhat less soluble than their native counterparts.
Crystallization and Data Collection
Crystals of native protein were grown by hanging drop vapor diffusion method at 4°C. One to 2 μl of protein solution were mixed with an equal amount of well solution (20%–25% PEG 2K monomethyl ether, 100 mM Na+·HEPES at pH 7.2–8.4, 0–200 mM NaCl) on a siliconized glass coverslip and equilibrated against 1 ml of the well solution. Crystals appeared within 3 days and grew to a maximum size of 1.0 mm × 0.4 mm × 0.4 mm in a week. The crystals were harvested in 30% PEG 2K monomethyl ether, 100 mM Na+·HEPES (pH 7.6), 20 mM Tris·Cl (pH 7.5), 120 mM NaCl, and 25%–30% ethylene glycol as cryoprotectant, then flash frozen in liquid nitrogen–cooled liquid propane and stored in liquid nitrogen. The crystals belong to space group P212121 with cell constants a = 58.12 Å, b = 87.50 Å, c = 117.66 Å. The calculated Matthew’s coefficient of 2.27 Å3/Dalton is consistent with two dimer molecules per asymmetric unit. Se-Met crystals were obtained and harvested in the same way as the native crystals except that the optimal conditions for Se-Met crystal growth require less concentrated protein (10 mg/ml instead of 14 mg/ml) and lower precipitant concentration in the well solution (16%–17% instead of 20%–25% PEG 2K monomethyl ether) in the drop. The Se-Met crystals are isomorphous to the native crystals (Table 1).
MAD data sets were measured at 100 K at beamline 5.0.2 of the Advanced Light Source (ALS). An ADSC Quantum 4 CCD detector was used to record the diffraction data in 1° oscillation steps at 10 s/frame over a 95° range. Data were measured at four wavelengths: low-energy remote (0.9808 Å), inflection (0.9801 Å), peak (0.9799 Å), and high-energy remote (0.9500 Å). At each wavelength, Friedel mates were measured by rotating the crystal 180° from φinit and measuring oscillation data over a second 95° range. A native data set was also measured at λ = 0.9810 Å using 20 s exposures. Diffraction data were reduced using the HKL software package (Otwinowski, 1993) (Table 1). The MAD data sets were scaled but not merged (option “no merge original index” in SCALEPACK). Local scaling was performed within SOLVE v. 1.14 as part of the phasing procedure (Terwilliger and Berendzen, 1999).
Structure Determination and Model Refinement
The automated structure determination program SOLVE version 1.14 (Terwilliger and Berendzen, 1999) was used to locate selenium atoms and compute MAD phases. The anomalous scattering factors f′ and f″ were obtained from experimental fluorescence spectra using the program KRAMIG (D. T. Cromer, Los Alamos National Laboratory, unpublished program). The low-energy remote wavelength data set was used as pseudonative data set. Thirteen out of 20 selenium sites in the asymmetric unit were found, and MAD phasing computations were carried out automatically using SOLVE v 1.14 with all four MAD data sets. MAD phasing power and figure of merit (FOM) were computed with CNS_SOLVE v. 0.5 (Brünger et al., 1998) (Table 1). Program DM (Collaborative Computational Project, 1994) was used to refine phases by solvent flattening (with estimated 40% solvent content) and histogram matching, thereby increasing the FOM to 0.80. The resulting map was of excellent quality, allowing most of the main chain and side chain atoms to be located. The seven selenium atoms not found by SOLVE are located in the N and C termini of Tube-DD and Pelle-DD for which no density is discernible. Model building was performed using the program O version 6.1.0 (Jones and Kjeldgaard, 1993).
The program CNS_SOLVE 0.5 was used to refine the model against the native data set. During each round of refinement, positional refinement by Powell minimization, individual B factor refinement, and slow-cool simulated annealing molecular dynamics were carried out along with anisotropic B factor correction, bulk solvent correction, and maximum likelihood refinement target using amplitudes as implemented in the CNS_SOLVE 0.5 program. Significant structural differences between the two copies of Tube-DD and Pelle-DD in the asymmetric unit precluded the use of noncrystallographic symmetry restraints. Sigma A weighted 2Fo – Fc and Fo – Fc maps were computed for visual inspection and model rebuilding. Water molecules included in the model form at least one hydrogen bond with protein atoms or other water oxygens atoms with B factors below 80 Å2. Stereochemical tests by PROCHECK (Laskowski et al., 1993) indicated that 93.7% of the residues fall in the most favorable regions and no residues fall in disallowed regions of the Ramachandran plot (Ramachandran and Sassiekharan, 1968). Coordinates have been deposited in the Protein Data Bank (Bernstein et al., 1977) with ID code 1d2z.
Site-Directed Mutagenesis, RNA Synthesis, Embryo Injection, and Cuticle Preparations
Mutant forms of Tube and Pelle were obtained through PCR amplification from cDNAs, using the method of Ho et al. (1989). The sequences of primers used are available upon request. Approximately 200 ng of PCR product was used as templates in RNA synthesis reactions utilizing the Ambion Megascript Sp6 kit supplemented with cap analog. Each RNA was suspended at a concentration of 1 mg/ml in injection buffer (2.5 mM PIPES [pH 6.9], 25 mM KCl, 0.5 mM EDTA, 25% glycerol). The recipient embryos were obtained from maternal flies of genotype tub[R5.6]/Df(3R)XM3 in the case of Tube construct injections, a genotype that produces no Tube transcript or protein (Letsou et al., 1993). In the case of Pelle construct injections, the genotype of the maternal flies was pll[25]/Df(3R)IR16, a genotype that produces no functional Pelle protein (Hecht and Anderson, 1993; P. T., unpublished data). Embryos were collected and microinjected as previously described (Anderson and Nüsslein-Volhard, 1984) and were later prepared and scored for cuticular markers as described (Wieschaus and Nüsslein-Volhard, 1986).
Protein Sample Preparation and Immunoblot Analysis
Lysates were prepared from injected embryos as described (Reach et al., 1996). Immunoblot analysis was done as described (Gillespie and Wasserman, 1994), with the primary antibodies used being a rabbit anti-Tube polyclonal sera (Letsou et al., 1993) or a rabbit anti-Pelle polyclonal sera, raised against amino acids 1–129 of Pelle.
Acknowledgments
We thank Zhiping Liu for her enthusiastic contributions throughout the stages of protein expression and characterization; John J. G. Tesmer for his assistance with data collection and MAD data analysis in addition to his guidance in structure refinement; David E. Coleman for enlightening discussions; Ryan Ulaszek for assistance with site-specific mutagenesis; Lothar Esser with figure preparation; and the staff of the ALS. This work is supported by National Institutes of Health grant GM50545 to S. A. W. and Welch Foundation grant I1229 to S. R. S.
Footnotes
Protein Data Bank ID Code
The coordinates and structure factors have been deposited with the ID code 1d2z.
References
- Anderson KV, Nüsslein-Volhard C. Information for the dorsal-ventral pattern of the Drosophila embryo is stored as maternal mRNA. Nature. 1984;311:223–227. doi: 10.1038/311223a0. [DOI] [PubMed] [Google Scholar]
- Baker SJ, Reddy EP. Modulation of life and death by the TNF receptor superfamily. Oncogene. 1998;17:3261–3270. doi: 10.1038/sj.onc.1202568. [DOI] [PubMed] [Google Scholar]
- Belvin MP, Jin Y, Anderson KV. Cactus protein degradation mediates Drosophila dorsal-ventral signaling. Gene Dev. 1995;9:783–793. doi: 10.1101/gad.9.7.783. [DOI] [PubMed] [Google Scholar]
- Bernstein FC, Koetzle TF, Williams JB, Meyer EF, Jr., Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M. The protein databank: a computer-based archival file for macromolecular structure. J. Mol. Biol. 1977;112:535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Boldin MP, Mett IL, Varfolomeev EE, Chumakov I, Shemer-Avni Y, Camonis JH, Wallach D. Self-association of the “death domains” of the p55 tumor necrosis factor (TNF) receptor and Fas/APO1 prompts signaling for TNF and Fas/APO1 effects. J. Biol. Chem. 1995;270:387–391. doi: 10.1074/jbc.270.1.387. [DOI] [PubMed] [Google Scholar]
- Brünger AT. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, Gros P, Grosse-Kunstleve RW, Jiang J-S, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography and NMR system (CNS): a new software suite for macromolecular structure determination. Acta Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Chang HY, Yang X, Baltimore D. Dissecting Fas signaling with an altered-specificity death-domain mutant: requirement of FADD binding for apoptosis but not Jun N-terminal kinase activation. Proc. Natl. Acad. Sci. USA. 1999;96:1252–1256. doi: 10.1073/pnas.96.4.1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou JJ, Matsuo H, Duan H, Wagner G. Solution structure of the RAIDD CARD and model for CARD/CARD interaction in caspase-2 and caspase-9 recruitment. Cell. 1998;94:171–180. doi: 10.1016/s0092-8674(00)81417-8. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, No. 4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Eberstadt M, Huang B, Chen Z, Meadows RP, Ng SC, Zheng L, Lenardo MJ, Fesik SW. NMR structure and mutagenesis of the FADD (Mort1) death-effector domain. Nature. 1998;392:941–945. doi: 10.1038/31972. [DOI] [PubMed] [Google Scholar]
- Edwards DN, Towb P, Wasserman SA. An activity-dependent network of interactions links the rel protein Dorsal with its cytoplasmic regulators. Development. 1997;124:3855–3864. doi: 10.1242/dev.124.19.3855. [DOI] [PubMed] [Google Scholar]
- Esnouf RM. An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J. Mol. Graph. 1997;15:133–138. doi: 10.1016/S1093-3263(97)00021-1. [DOI] [PubMed] [Google Scholar]
- Esser L. Gl_render version 0.7. 1999 http://www.hhmi.swmed.edu/external/Doc/Gl_render/Html/gl_render.html.
- Feinstein E, Kimchi A, Wallach D, Boldin M, Varfolomeev E. The death domain: a module shared by proteins with diverse cellular functions. Trends Biochem. Sci. 1995;20:342–344. doi: 10.1016/s0968-0004(00)89070-2. [DOI] [PubMed] [Google Scholar]
- Galindo RL, Edwards DN, Gillespie SK, Wasserman SA. Interaction of the pelle kinase with the membrane-associated protein tube is required for transduction of the dorsoventral signal in Drosophila embryos. Development. 1995;121:2209–2218. doi: 10.1242/dev.121.7.2209. [DOI] [PubMed] [Google Scholar]
- Gillespie SK, Wasserman SA. Dorsal, a Drosophila Rel-like protein, is phosphorylated upon activation of the transmembrane protein Toll. Mol. Cell. Biol. 1994;14:3559–3568. doi: 10.1128/mcb.14.6.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Großhans J, Bergmann A, Haffter P, Nüsslein-Volhard C. Activation of the kinase Pelle by Tube in the dorsoventral signal transduction pathway of Drosophila embryo. Nature. 1994;372:563–566. doi: 10.1038/372563a0. [DOI] [PubMed] [Google Scholar]
- Großhans J, Schnorrer F, Nüsslein-Volhard C. Oligomerization of Tube and Pelle leads to nuclear localisation of dorsal. Mech. Dev. 1999;81:127–138. doi: 10.1016/s0925-4773(98)00236-6. [DOI] [PubMed] [Google Scholar]
- Hecht PM, Anderson KV. Genetic characterization of tube and pelle, genes required for signaling between Toll and dorsal in the specification of the dorsal-ventral pattern of the Drosophila embyro. Genetics. 1993;135:407–417. doi: 10.1093/genetics/135.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson WA. Determination of macromolecular structures from anomalous diffraction of synchrotron radiation. Science. 1991;254:51–58. doi: 10.1126/science.1925561. [DOI] [PubMed] [Google Scholar]
- Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- Hoffmann JA, Kafatos FC, Janeway CA, Ezekowitz RA. Phylogenetic perspectives in innate immunity. Science. 1999;284:1313–1318. doi: 10.1126/science.284.5418.1313. [DOI] [PubMed] [Google Scholar]
- Huang B, Eberstadt M, Olejniczak ET, Meadows RP, Fesik SW. NMR structure and mutagenesis of the Fas (APO-1/CD95) death domain. Nature. 1996;384:638–641. doi: 10.1038/384638a0. [DOI] [PubMed] [Google Scholar]
- Jeong EJ, Bang S, Lee TH, Park YI, Sim WS, Kim KS. The solution structure of FADD death domain. Structural basis of death domain interactions of Fas and FADD. J. Biol. Chem. 1999;274:16337–16342. doi: 10.1074/jbc.274.23.16337. [DOI] [PubMed] [Google Scholar]
- Jones TA, Kjeldgaard M. O Version 5.9. Second Edition Uppsala University; Uppsala, Sweden: 1993. [Google Scholar]
- Kraulis PJ. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 1991;24:946–950. [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. The dorsoventral regulatory gene cassette spätzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86:973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- Leszczynski JF, Rose GD. Loops in globular proteins: a novel category of secondary structure. Science. 1986;234:849–855. doi: 10.1126/science.3775366. [DOI] [PubMed] [Google Scholar]
- Letsou A, Alexander S, Orth K, Wasserman SA. Genetic and molecular characterization of tube, a Drosophila gene maternally required for embryonic dorsoventral polarity. Proc. Natl. Acad. Sci. USA. 1991;88:810–814. doi: 10.1073/pnas.88.3.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letsou A, Alexander S, Wasserman SA. Domain mapping of tube, a protein essential for dorsoventral patterning of the Drosophila embryo. EMBO J. 1993;12:3449–3458. doi: 10.1002/j.1460-2075.1993.tb06019.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liepinsh E, Ilag LL, Otting G, Ibanez CF. NMR structure of the death domain of the p75 neurotrophin receptor. EMBO J. 1997;16:4999–5005. doi: 10.1093/emboj/16.16.4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R, Janeway CA., Jr. An ancient system of host defense. Curr. Opin. Immunol. 1998;10:12–15. doi: 10.1016/s0952-7915(98)80024-1. [DOI] [PubMed] [Google Scholar]
- Medzhitov R, Preston-Hurlburt P, Kopp E, Stadlen A, Chen C, Ghosh S, Janeway CA., Jr. MyD88 is an adaptor protein in the hToll/IL-1 receptor family signaling pathways. Mol. Cell. 1998;2:253–258. doi: 10.1016/s1097-2765(00)80136-7. [DOI] [PubMed] [Google Scholar]
- Muzio M, Ni J, Feng P, Dixit VM. IRAK (Pelle) family member IRAK-2 and MyD88 as proximal mediators of IL-1 signaling. Science. 1997;278:1612–1615. doi: 10.1126/science.278.5343.1612. [DOI] [PubMed] [Google Scholar]
- Nicholls A, Sharp KA, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins: Struct. Funct. Genet. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. Oscillation data reduction program. In: Sawyer NIL, Bailey SW, editors. Data Collection and Processing. Science and Engineering Council Daresbury Laboratory; Daresbury, U.K.: 1993. pp. 56–62. [Google Scholar]
- Park A, Baichwal VR. Systematic mutational analysis of the death domain of the tumor necrosis factor receptor 1-associated protein TRADD. J. Biol. Chem. 1996;271:9858–9862. doi: 10.1074/jbc.271.16.9858. [DOI] [PubMed] [Google Scholar]
- POV-ray team POV-ray: the persistence of vision ray tracer. 1998 http://www.povray.org/
- Qin H, Srinivasula SM, Wu G, Fernandes-Alnemri T, Alnemri ES, Shi Y. Structural basis of procaspase-9 recruitment by the apoptotic protease-activating factor 1. Nature. 1999;399:549–557. doi: 10.1038/21124. [DOI] [PubMed] [Google Scholar]
- Ramachandran GN, Sassiekharan V. Conformation of polypeptides and proteins. Adv. Protein Chem. 1968;28:283–437. doi: 10.1016/s0065-3233(08)60402-7. [DOI] [PubMed] [Google Scholar]
- Reach M, Galindo RL, Towb P, Allen JL, Karin M, Wasserman SA. A gradient of cactus protein degradation establishes dorsoventral polarity in the Drosophila embryo. Dev. Biol. 1996;180:353–364. doi: 10.1006/dbio.1996.0308. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1989. [Google Scholar]
- Shelton CA, Wasserman SA. Pelle encodes a protein kinase required to establish dorsoventral polarity in the Drosophila embryo. Cell. 1993;72:515–525. doi: 10.1016/0092-8674(93)90071-w. [DOI] [PubMed] [Google Scholar]
- Tartaglia LA, Ayres TM, Wong GHW, Goeddel DV. A novel domain within the 55 kd TNF receptor signals cell death. Cell. 1993;74:845–853. doi: 10.1016/0092-8674(93)90464-2. [DOI] [PubMed] [Google Scholar]
- Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr. D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towb P, Galindo RL, Wasserman SA. Recruitment of Tube and Pelle to signaling sites at the surface of the Drosophila embryo. Development. 1998;125:2443–2450. doi: 10.1242/dev.125.13.2443. [DOI] [PubMed] [Google Scholar]
- Wieschaus E, Nüsslein-Volhard C. Looking at embryos. In: Roberts DB, editor. Drosophila: A Practical Approach. IRL Press; Oxford: 1986. pp. 199–227. [Google Scholar]
- Yang W, Hendrickson WA, Kalman ET, Crouch RJ. Expression, purification, and crystallization of natural and selenomethionyl recombinant ribonuclease H from Escherichia coli. J. Biol. Chem. 1990;265:13553–13559. [PubMed] [Google Scholar]