Summary
MicroRNAs (miRNAs) regulate target mRNAs through a combination of translational repression and mRNA destabilization, with mRNA destabilization dominating at steady state in the few contexts examined globally. Here, we extend the global steady-state measurements to additional mammalian contexts and find that regardless of the miRNA, cell type, growth condition, or translational state, mRNA destabilization explains most (66% to >90%) miRNA-mediated repression. We also determine the relative dynamics of translational repression and mRNA destabilization for endogenous mRNAs as a miRNA is induced. Although translational repression occurs rapidly, its effect is relatively weak, such that by the time consequential repression ensues, the effect of mRNA destabilization dominates. These results imply that consequential miRNA-mediated repression is largely irreversible and provide other insights into the nature of miRNA-mediated regulation. They also simplify future studies, dramatically extending the known contexts and time points for which monitoring mRNA changes captures most of the direct miRNA effects.
Introduction
MicroRNAs are small, non-coding RNAs that post-transcriptionally regulate the expression of most mammalian genes (Bartel, 2009; Friedman et al., 2009). Acting as the specificity components of ribonucleoprotein silencing complexes, miRNAs pair with target mRNAs at sites complementary to the miRNA 5′ region. Most effective sites map to 3′ untranslated regions (3′ UTRs) and pair perfectly with the miRNA seed (nucleotides 2–7), with an additional pair at nucleotide 8 and/or an A across from nucleotide 1 (Bartel, 2009).
Although early reports of gene regulation by miRNAs emphasized their role as translational repressors (Wightman et al., 1993; Olsen and Ambros, 1999; Seggerson et al., 2002), subsequent studies revealed that miRNAs can also induce mRNA degradation (Bagga et al., 2005; Krutzfeldt et al., 2005; Lim et al., 2005). This degradation is a consequence of miRNA-mediated deadenylation of target mRNAs (Behm-Ansmant et al., 2006; Giraldez et al., 2006; Wu et al., 2006), which causes these mRNAs to undergo decapping and then 5′–3′ decay (Rehwinkel et al., 2005; Behm-Ansmant et al., 2006; Chen et al., 2009). The discovery of this second mode of repression raised the question as to the relative contributions of translational repression and mRNA degradation to reducing the protein abundance of regulated genes.
Large-scale analyses comparing protein and mRNA changes of predicted miRNA targets after introducing or deleting individual mammalian miRNAs found that protein changes generally correspond to changes in polyadenylated mRNA abundance (Baek et al., 2008). More precise measurements comparing changes in translational efficiency (TE) to changes in mRNA again found that mRNA degradation explains the majority of miRNA-mediated repression, with translational repression contributing roughly 10–25% of the overall repression (Hendrickson et al., 2009; Guo et al., 2010). These global measurements of TE and mRNA (or protein and mRNA) were made at relatively late time points (12–32 hours after introducing the miRNA or long after induction of an endogenous miRNA), and thus are thought to reflect the steady-state effects of the miRNA (Baek et al., 2008; Hendrickson et al., 2009; Guo et al., 2010). When miRNAs are expressed at constant levels, steady-state measurements are ideal for quantifying the relative contributions of translational repression and mRNA degradation because they integrate effects occurring throughout the life cycle of each targeted transcript.
If generalizable to other cell types and conditions, these high-throughput steady-state measurements, which indicate that mRNA changes closely approximate the overall effects of a miRNA on target gene expression, would be welcome news for those placing mammalian miRNAs into gene-regulatory networks and quantifying their impact on gene expression, since measuring changes in mRNA levels is much easier than measuring changes in protein levels or TE. However, protein/TE and mRNA effects have been globally compared in only two cell lines, HeLa cells (Baek et al., 2008; Selbach et al., 2008; Guo et al., 2010) and HEK293T cells (Hendrickson et al., 2009), and a single primary cell type, mouse neutrophils (Baek et al., 2008; Guo et al., 2010), which leaves open the possibility that translational repression might dominate in most other mammalian contexts.
The observation that mRNA destabilization can account for most repression at steady state has prompted a search for time points in which translational repression might explain a larger proportion of the repression. Two studies examined the dynamics of miRNA-mediated repression on inducible reporter genes as these genes begin to be expressed in fly and human cells (Bethune et al., 2012; Djuranovic et al., 2012), and another examined the effects of miR-430 on its endogenous targets in the zebrafish embryo (Bazzini et al., 2012). In blastula-stage zebrafish embryos (4 hours postfertilization [hpf]), miR-430 substantially reduces the TE of its targets with little effect on their stability, whereas by gastrulation (6 hpf), the relative contributions of TE and mRNA destabilization closely resemble those observed previously at steady state in mammalian systems (Baek et al., 2008; Hendrickson et al., 2009; Guo et al., 2010; Bazzini et al., 2012). Because miR-430 is strongly induced shortly before the blastula stage, the large amount of translational repression observed in the blastula stage, followed by the mRNA destabilization observed later in the gastrula stage, was proposed to reflect the fundamental dynamics of miRNA-mediated repression (Bazzini et al., 2012).
The idea that miRNA-mediated translational repression precedes mRNA degradation cannot be disputed—an mRNA molecule can undergo translational repression only before it has been degraded and thus its translational regulation must precede regulation at the level of its stability in the same way that transcriptional regulation must precede translational regulation. However, subsequent insight into the shift in regulatory regime occurring as zebrafish embryos progress from pre- to post-gastrulation has overturned the idea that the miR-430 observations reflect the dynamics of miRNA-mediated repression (Subtelny et al., 2014). Prior to gastrulation, mRNA poly(A)-tail length and TE are coupled, and short-tailed mRNAs are stable. These two unique conditions enable miRNA-mediated deadenylation to cause translational repression without mRNA destabilization (Subtelny et al., 2014). The transition to mostly mRNA decay is due to a change in these conditions at gastrulation, such that coupling between tail length and TE is lost and short-tailed mRNAs become less stable, which causes the consequence of miRNA-mediated deadenylation to shift from translational repression to mRNA destabilization (Subtelny et al., 2014). When considering this shifting regulatory regime, the miR-430 results do not provide insight into the dynamics of the two modes of miRNA-mediated repression for endogenous mRNAs, nor do they demonstrate that miRNA-mediated translational repression occurring through a deadenylation-independent mechanism ever mediates meaningful changes in the expression of endogenous mRNAs. This being said, the miR-430 study is notable as the first (and only) to identify an endogenous setting in which the effects of a miRNA cannot be approximated by changes in mRNA levels (Bazzini et al., 2012). Because of the regulatory regime operating in the pre-gastrulation zebrafish embryo (and presumably in other early embryos or other unusual settings, such as neuronal synapses), measuring mRNA changes misses essentially all of the effects of miRNAs in this setting (Subtelny et al., 2014).
The two studies that monitor reporter genes rather than endogenous transcripts to examine miRNA repression dynamics both report that a phase of substantial translational repression occurs prior to detectable mRNA deadenylation or decay (Bethune et al., 2012; Djuranovic et al., 2012). However, the updated understanding of the miR-430 results reopens the question of whether such a phase also occurs for endogenous mRNAs. Although reporters can faithfully represent endogenous genes, several observations led us to suspect that when measuring the effects of miRNAs there might be a difference between reporters and endogenous genes. First, even at very early time points in zebrafish embryonic development, most repression of endogenous mRNAs is attributable to miRNA-mediated deadenylation rather than direct translational repression (Subtelny et al., 2014). Second, at steady state, the fractional repression attributed to translational repression of the reporters (Bethune et al., 2012; Djuranovic et al., 2012) exceeds that typically observed for endogenous mRNAs in mammalian cells (Baek et al., 2008; Hendrickson et al., 2009; Guo et al., 2010). Similarly, the magnitude of repression observed for reporters vastly exceeds that typically observed for endogenous mRNAs in mammalian cells.
Here we substantially expand the contexts and conditions for which the repressive effects on endogenous mRNAs are examined. We measured the consequence of deleting specific miRNAs on the mRNA and translation (or protein) of predicted targets in mouse liver, primary macrophages, and activated and non-activated primary B cells, thereby adding four additional biological settings to the previous two settings (mouse neutrophils and zebrafish embryos) in which translational effects on endogenous targets have been broadly measured. We also measured the translational effects on endogenous mRNAs after adding specific miRNAs in two additional cell lines (U2OS cells and NIH3T3 cells) and two additional conditions (growth-arrested cells and translationally inhibited cells). In all cases, mammalian miRNAs predominantly acted to decrease target mRNA levels, with relatively small contributions from translational repression. We then examined the repression dynamics of endogenous mRNAs and did not observe an early phase in which dominant translation effects imparted substantial repression. We conclude that although translational repression is rapid, its effect is relatively weak, and thus by the time consequential repression ensues, the effect of mRNA destabilization dominates.
Results
Negligible contribution of nuclear or deadenylated RNA to TE changes
The adaptation of ribosome profiling to mammalian cells has provided a sensitive and quantitative method to assess the influence of miRNAs on TE (Guo et al., 2010). Ribosome profiling uses high-throughput sequencing of ribosome-protected fragments (RPFs) to determine the positions of millions of ribosomes on mRNAs (Ingolia et al., 2009). To assess the TE of a gene, RPFs mapping to its open reading frame are normalized to its mRNA abundance, as determined by RNA-seq.
When comparing samples with and without a particular miRNA, the change in RPFs for a target of that miRNA reflects the aggregate effects of mRNA degradation and translational repression, while the change in mRNA reflects only the component attributable to degradation. After accounting for the change in RPFs attributed to mRNA degradation, the residual change in RPFs reflects a change in TE, which is interpreted as the miRNA-mediated translational repression acting on the message at the moment the ribosomes were arrested.
Previously, we observed little miRNA-mediated translational repression in mammalian cells, with the concern that these modest TE changes might actually be overestimates (Guo et al., 2010). An overestimation would occur if some polyadenylated mRNA were sequestered away from the compartment containing both miRNAs and ribosomes, as would be the case for mRNAs awaiting export from the nucleus. In this case, miRNA-mediated degradation of mRNAs only in the cytoplasm would lead to a larger relative loss of RPFs (which are only from the cytoplasm) than mRNA fragments (which are from both the nucleus and cytoplasm), thereby inflating the apparent translational repression. To address this concern, we performed ribosome profiling on miRNA- and mock-transfected U2OS cells, and in parallel performed RNA-seq on poly(A)-selected RNA from both whole-cell lysates and cytoplasmic fractions. The efficacy of fractionation was demonstrated by the depletion of pre-ribosomal RNAs (pre-rRNAs) in the cytoplasmic fraction (Figure S1A). Following transfection of miR-1, a miRNA not normally expressed in U2OS cells, repression was observed, with significant degradation of mRNAs with at least one miR-1 3′-UTR site (Figure 1A). The amount of degradation was indistinguishable in the RNA-seq libraries made with either whole-cell or cytoplasmic mRNA, and thus the amount of translational repression was similarly indistinguishable (Figure 1A). The same was observed with miR-155, another miRNA not normally expressed in U2OS cells, demonstrating that a nuclear mRNA-sequestration artifact does not detectably elevate the signal for miRNA-mediated translational repression in mammalian cells.
Figure 1. Steady-State Changes in Gene Expression Due to miRNAs.
(A) The influence of using different types of mRNA enrichment when measuring the effects of miRNAs on mRNA levels and TE. Plots show cumulative distributions of changes in RPFs (top), mRNA (middle), and TE (bottom) after transfection of either miR-1 (left) or miR-155 (right) into U2OS cells. The impact of the miRNA on genes with at least one site to the cognate miRNA in their 3′ UTR (≥1 site; n = 1321 and 1075 for miR-1 and miR-155, respectively) is compared to that of control genes (no site; n = 1205 and 1056, respectively), which were chosen from the genes with no site to the cognate miRNA throughout their entire transcript to match the 3′-UTR-length distributions of site-containing genes. The three types of mRNA enrichment were poly(A)-selected total RNA, poly(A)-selected cytoplasmic RNA, and tRNA/rRNA-depleted total RNA [total p(A), cyto p(A), and Ribo-zero, respectively]. RNA-seq analyses of these preparations were used to calculate mRNA and TE changes, with results plotted as indicated in the key. Data were normalized to the median changes observed for the controls. See also, Figure S1.
(B) A simplified representation of the results in panel A showing for each experiment the mean RPF fold change (log2) attributable to changes in mRNA (blue) and TE (green), after subtracting the mean RPF change of the no-site control genes. The bars for the percent contribution attributable to mRNA and TE changes are calculated using the mean RNA and RPF fold changes (log2) after normalizing to the median no-site fold change (log2) (Figure S2). The schematic (left) depicts the components of the compound bar graphs (right). Significant changes for each component are indicated with asterisks of the corresponding color [*, P ≤0.05; **, P ≤0.001, one-tailed Kolmogorov–Smirnov test (K–S test)], with the relative contribution of TE to repression (Figure S2D) reported as a percentage in green below each bar. See also, Figure S2.
(C) The steady-state effects of miRNAs in a variety of cell types, shown using compound bar graphs like those of panel B. For comparison with our current results, previously published results from HeLa and neutrophils (neut.) (Guo et al., 2010) are also plotted after reanalysis using the current methods (including the method for choosing no-site control cohorts). When available, proteomics-supported predicted targets were also analyzed (right). For HeLa and neutrophil these were the ones selected previously (Guo et al., 2010), and for the other samples these were selected from our proteomics data as the subset of site-containing genes with fold changes (log2) ≤−0.3 in the presence of the miRNA. Experiments with cell lines compared cells with and without the miRNA introduced by either transfection (HeLa and 293T) or induction from a transgene (3T3). Experiments with B cells, neutrophils and liver compared cells/tissues isolated from wild-type and miRNA-knockout mice. The hours indicate the time following transfection (HeLa and 293T), induction (3T3), or activation (B cells). See also, Figure S3, Table S1 and Table S2.
(D) Comparison of mRNA and RPF changes for individual genes analyzed in panels (A)–(C). For U2OS cells, the results for the poly(A)-selected cytoplasmic RNA are shown. The dashed line is for y = x; the vertical and horizontal lines indicate the mean fold changes for the correspondingly colored groups of genes. Red, genes with ≥1 3′-UTR site to the cognate miRNA; grey, no site to the miRNA selected as in panel A; green, proteomics-supported predicted targets (Table S1 and Table S2). Data were normalized to the median changes observed for the controls. A comparable analysis of the HeLa and neutrophil data has been published (Guo et al., 2010).
A second concern involved the measurement of poly(A)-selected RNA. Monitoring changes in poly(A)-selected RNA leaves unanswered the question of whether repressed mRNAs are degraded or merely deadenylated, and under-recovery of partially deadenylated messages during poly(A)-selection might overestimate the amount of mRNA degradation that has occurred. To address this concern, we generated a third set of RNA-seq libraries from the aforementioned U2OS cells, starting with whole-cell RNA preparations that were not poly(A)-selected and instead were depleted of both tRNAs and rRNAs. Greatly increased RNA-seq coverage of replication-dependent histone mRNAs, which lack poly(A) tails, illustrated our ability to detect RNAs regardless of poly(A)-tail length (Figure S1B). Results for miRNA-dependent changes in tRNA/rRNA-depleted RNA were similar to those of poly(A)-selected RNA (Figure 1A), which indicated that changes in accumulation of mRNA refractory to poly(A) selection were negligible. These results imply that the absolute amount of deadenylated mRNAs and other intermediates underrepresented in poly(A)-selected RNA is small, even for repressed mRNAs, presumably because these decay intermediates are rapidly decapped and degraded. Thus, concerns that translational repression measurements might have been either under- or over-estimates appear to be unfounded; comparing TEs calculated by simply normalizing RPF changes to those of poly(A)-selected RNA accurately measures translational repression in mammalian cells.
To aid comparisons, the results in Figure 1A can be summarized in compound bar graphs (Figure 1B). For each experiment, the mean RPF fold change (distance that the compound bar extends below zero) indicates the overall repression. The mRNA contribution (blue component of the compound bar) indicates the extent to which mRNA degradation explains this repression, and any residual RPF change is the TE contribution (green component), which reflects the translational repression of the remaining mRNA. Based on the RPF reductions attributable to these two repression modes, their relative contributions to repression are then calculated (Figure S2). Of the two modes, mRNA degradation dominates in U2OS cells (Figure 1B), despite the presence of a P-body subtype reported to impart increased translational repression (Castilla-Llorente et al., 2012).
Dominant mRNA destabilization in many contexts
We expanded our analysis to examine the steady-state effects of gaining or losing a miRNA in additional cell lines and biological contexts. These experiments included studies comparing RPF and mRNA measurements in liver from wild-type mice, which expresses miR-122, to those in liver from mice lacking the mir-122 gene. Similarly, the effects of miR-155 in activated primary murine B cells were measured comparing cells from wild-type mice to those lacking the mir-155 gene. These loss-of-function experiments enabled analysis of endogenous targets in their endogenous settings. The effects on predicted targets of endogenous miR-122 in mouse liver, endogenous miR-155 in primary mouse B cells, induced miR-1 (expressed from a transgene) in 3T3 cells, and transfected miR-1 in HEK293T cells all resembled the published effects of endogenous miR-223 in neutrophils and transfected miRNAs in HeLa cells (Figure 1C). In all settings, reduced mRNA levels explained most of the steady-state RPF reduction observed in the presence of the miRNA, implying that miRNAs predominantly act to reduce target mRNA levels. Nonetheless, mean RPF reduction attributable to translational repression was observed, ranging from 1–34% of the total, depending on the experiment.
Because a 7–8-nt site to a miRNA is not always sufficient to mediate miRNA targeting, high-throughput proteomic measurements can be used to identify high-confidence targets by identifying site-containing genes with less protein in the presence of the miRNA (Guo et al., 2010). With this in mind, we performed a quantitative proteomics experiment using SILAC (stable isotope labeling with amino acids in culture) to identify a set of genes with reduced protein after inducing miR-1 in 3T3 cells (Table S1) and pulsed SILAC (Selbach et al., 2008) to identify those responding to miR-155 in activated B cells (Table S2). These proteomics-supported predicted targets showed greater mean repression than did the complete set of genes with ≥1 site, as expected if they were enriched in direct targets of the miRNA (Figure 2C). For new and published experiments with proteomics-supported predicted targets, the fractional repression attributed to translational repression ranged from 6–26%, somewhat narrower than the range observed when considering all mRNAs with sites, perhaps because a focus on the more confidently identified targets decreased experimental variability.
Figure 2. Minor Impact of Translational Repression at all Times in Mammalian Cells.
(A) Induction of miRNAs in activated murine B cells and in contact-inhibited NIH3T3 cells engineered to inducibly express miR-1, miR-124, or miR-155. Induction was monitored using RNA blots, probing for the induced miRNA. For samples from B cells, the membrane was reprobed for endogenous U6 snRNA, which served as a loading control for normalization, and expression is plotted relative to that of the non-activated cells. For samples from 3T3 cells, synthetic standards for the induced miRNAs and endogenous miR-21 were included on the blot, and used for absolute quantification. Expression is plotted relative to that of miR-21, with relative expression of the let-7 family (inferred from small-RNA sequencing data) also shown.
(B) The contributions of mRNA decay and translational repression following miR-155 induction in primary murine B cells. The same sets of site-containing and control genes are analyzed in all time points. If the contribution of TE was calculated to be less than 0, the value reported below the bar was 0. Otherwise, as in Figure 1C. The 48 h time point is replotted from Figure 1C and was from a preparation of cultured B cells independent from that used for the earlier time points. See also, Table S2.
(C) The contributions of mRNA decay and translational repression following induction of miR-155 (top), miR-1 (middle), or miR-124 (bottom) in the corresponding contact-inhibited 3T3 cell lines. In the absence of proteomics data for miR-124, the top 100 site-containing genes, as ranked by total context+ score (Garcia et al., 2011) regardless of site conservation, were analyzed to focus on a subset of site-containing genes likely to be regulated by miR-124. Otherwise, as in panel B. The miR-1 48 h time point is replotted from Figure 1C and is from the same experiment as the earlier time points. See also, Table S1 and Table S3.
Although the amount of repression attributed to translational repression did not always reach statistical significance, our results are consistent with the idea that a small amount of translational repression occurs for each direct target in each context. As was found previously (Baek et al., 2008; Hendrickson et al., 2009; Guo et al., 2010), a gene-by-gene analysis of results from each of the newly examined settings revealed no compelling evidence for a subset of genes repressed at only the translational level (Figure 1D), although the possibility of a few such genes cannot be ruled out.
Matching mRNA and proteomic results for less proficient miRNAs
In pilot experiments aimed at extending our studies to other endogenous contexts, we used wild-type and miRNA-deleted mice to acquire mRNA microarray data for macrophages and neutrophils with and without miR-21, and B cells with and without miR-150. Although these miRNAs were each among the most frequently sequenced miRNAs in the respective wild-type cells (Figure S3A), we observed weak miRNA effects when comparing sets of genes with and without 3′-UTR sites to the cognate miRNA (Figure S3B).
A potential explanation for the weak signals observed by mRNA profiling was that most of the repression was occurring through translational repression rather than mRNA degradation. However, when we used quantitative proteomics to test this possibility, the proteomics results mirrored those of the mRNA arrays, providing no evidence for substantial translational repression (Figure S3B and Table S3). Thus the weak repression signals observed at the mRNA level for endogenously expressed miR-21 and miR-150 were not due to a discrepancy between mRNA changes and the overall effects of miRNA-mediated repression. These results add to the growing list of endogenous settings for which mRNA changes accurately represent the effects of miRNA-mediated repression. This list now includes miR-223 in neutrophils (Baek et al., 2008; Guo et al., 2010), miR-21 in macrophages and neutrophils (Figure S3B), miR-122 in liver (Figure 1C), miR-150 in primary B cells (Figure S3B), and miR-155 in activated B cells (Figure 1C).
Dynamics of endogenous mRNA repression by inducible miRNAs
The shifting regulatory regime in the early zebrafish embryo, which changes the consequences of miRNA-mediated poly(A)-tail shortening, confounded the previous attempt to determine the dynamics of the two modes of repression for endogenous messages (Bazzini et al., 2012; Subtelny et al., 2014). Therefore, we set out to characterize the regulatory dynamics of miRNA-mediated repression of endogenous mRNAs and determine if there might be an endogenous setting in which these dynamics could give rise to a phase of substantial translation-dominated repression, as previously observed in reporter experiments (Bethune et al., 2012; Djuranovic et al., 2012).
Perhaps the most dynamic mammalian miRNA is miR-155, which is rapidly and strongly induced in B and T cells upon activation (Thai et al., 2007). In primary murine B cells, we observed a nearly 10-fold increase 4 h after activation with lipopolysaccharide, IL-4, and anti-CD40 (Figure 2A). Although presumably not as strong as for miR-430 in zebrafish embryos [which is expressed from as many as 93 loci (Giraldez et al., 2005)], miR-155 induction was nonetheless stronger than that of other mammalian miRNAs, in that no other mammalian miRNA has been reported to increase so rapidly to a high level of expression.
To assess the dynamics of translational repression and mRNA decay during miR-155–mediated repression, we isolated B cells from wild-type and miR-155 knockout mice, activated these cells, and then performed ribosome profiling and RNA-seq to monitor miRNA-dependent TE and mRNA changes occurring soon after induction. At 2 h post-activation, repression of genes with ≥1 site was detectable, but neither the mRNA nor the TE component was significantly decreased on its own. At 4 h, the small amount of repression was predominantly attributable to reduced TE (Figure 2B). By 8 h, the proportion attributed to translational repression abated, and at this time point the mRNA degradation so closely approached overall repression that the mean mRNA change for genes with ≥1 site slightly exceeded the mean RPF change (Figure 2B; P = 0.028, two-tailed K–S test for TE). Because this slight excess was not observed for the proteomics-supported predicted targets or in similar experiments with other miRNAs, we attribute it to experimental variability rather than evidence of translational activation. After 48 h, mRNA degradation continued to dominate (Figure 2B; as already shown in the steady-state analyses of Figure 1C), which indicated that B cells resemble other cells with respect to steady-state repression.
Although we found some evidence for translational repression dominating early in miR-155 induction, the amount of repression observed during this brief period was much weaker than that observed during the analogous phase of reporter experiments. Thus, we cannot claim to have found a mammalian setting with an early phase of substantial translational repression of endogenous messages, i.e., a time at which substantial repression would be missed if only mRNA changes were monitored. To further explore repression dynamics in mammalian cells, we created stable, miRNA-inducible 3T3 cell lines in which doxycycline treatment rapidly induced the expression of a miRNA not normally expressed in 3T3 cells (either miR-1, miR-124, or miR-155) to levels comparable to those of miR-21 and the let-7 miRNA family (Figure 2A), which are the miRNA and miRNA family most frequently sequenced for these cells (Rissland et al., 2011). The major advantage of such cell lines for studying the dynamics of translational repression and decay on endogenous messages is that, in contrast to B cells, miRNA induction does not accompany significant developmental changes, allowing the miRNA effects to be more easily isolated. With these lines we performed ribosome profiling and RNA-seq soon after miRNA induction, comparing translational efficiencies and mRNA expression levels with those of un-induced cells.
To account for the 2–3 h lag prior to the appearance of increased mature miRNA, the first time-point examined was 4 h post-induction. At 4 h, the miR-155–expressing line showed significant repression of genes with ≥1 site, all of which was attributed to translational repression (Figure 2C). At later time points, mRNA degradation dominated, as observed in B cells. For the miR-1–expressing line, 4 h was too early to observe significant repression for genes with ≥1 site, and by 8 h, mRNA degradation already dominated (Figure 2C), suggesting that we had missed any potential translation-dominant phase. For miR-124, a translation-dominant phase also was not observed (Figure 2C), presumably because induction was too gradual to achieve significant repression at early time points. (As we did not acquire murine proteomics data for miR-124, the top predicted targets were used instead of proteomics-supported targets.) Because miRNA induction in vivo is rarely more rapid than that achieved for miR-124 in our inducible line, we suggest that the miR-124 results are representative of most endogenous settings.
Minimal influence of translational stress and state
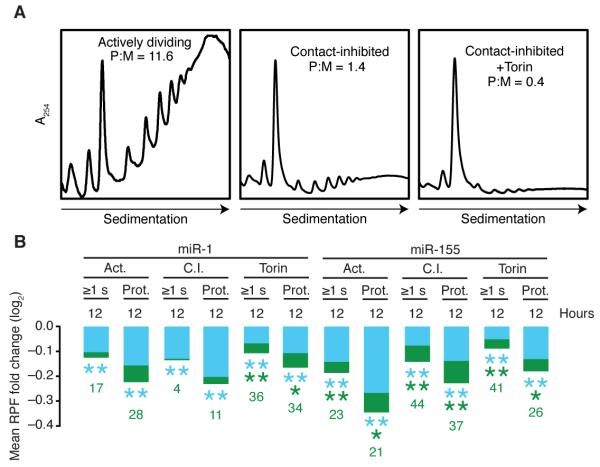
Having investigated eight different cell types and six different miRNAs, and having considered both pre-steady-state and later time points without identifying a setting with substantial overall repression in which translational effects dominated, we turned to the potential influence of cellular state. Studies of lin-4–mediated repression in C. elegans suggest that starvation might tip the balance towards more translational inhibition with less mRNA degradation (Holtz and Pasquinelli, 2009), presumably because starvation influences global translational activity. Therefore, we compared the relative contributions of TE and mRNA degradation for 3T3 cells in three translational states: 1) dividing cells, which have very active translation [polysome to monosome ratio (P:M) = 11.6], 2) contact-inhibited cells (P:M = 1.4), and 3) contact-inhibited cells under Torin1-induced mTOR inhibition (P:M = 0.4) (Figure 3A). We found no pervasive difference in the relative contribution of translational repression to miR-1– and miR-155–mediated repression between these states (Figure 3B), despite the ~30-fold range in translational activity. Thus, translational stress, and more generally the translational state, does not have a perceptible global impact on the mode of miRNA-mediated regulation in these mammalian cells.
Figure 3. Negligible Influence of Translational Stress or State on the Repression Mode.
(A) Polysome profiles showing the translational activity of actively dividing (left), contact-inhibited (middle), and Torin1-treated contact-inhibited (right) miR-1 inducible 3T3 cells. Profiles are normalized to the monosome peak, with the polysome-to-monosome ratio (P:M) indicated.
(B) The contributions of mRNA decay and translational repression following miR-1 or miR-155 induction in the corresponding 3T3 cell lines in the indicated states. Otherwise, as in Figure 2C. Results for contact-inhibited 3T3 cells expressing miR-1 and miR-155 were recalculated so as to only consider site-containing and no-site genes present in all samples. Abbreviations: Act., actively dividing; C.I., contact-inhibited; Torin, contact-inhibited and Torin1-treated; ≥1 s, genes with at least one site to the cognate miRNA in their 3′ UTR; Prot., proteomics-supported predicted targets. See also, Figure S4.
Because translating ribosomes displace miRNA-directed silencing complexes, which renders miRNA sites in the path of the ribosome much less effective than those ≥15 nt downstream of the stop codon (Grimson et al., 2007), we reasoned that the efficacy of sites within open reading frames (ORFs) might increase in conditions of reduced translational activity. Indeed, relative to the efficacy of 3′-UTR sites, the efficacy of ORF sites did appear to increase when translation was repressed with Torin1 (Figure S4A), which supported the model in which displacement of bound miRNAs by translating ribosomes is the predominant reason that ORF sites are ineffective.
Discussion
The principles of repression dynamics
Our results in 3T3 and B cells, considered in light of the fundamental differences between the nature of translational repression and mRNA destabilization, lead to the following principles regarding the miRNA-mediated repression of endogenous mRNAs in mammalian cells: Compared to translational repression, detectable mRNA destabilization occurs after more of a lag, presumably because mRNA decay takes longer than inhibiting translation initiation. Because of this relative lag, after unusually robust miRNA induction, we can detect a short phase resembling that observed in reporter experiments, in which most of the repression is from decreased TE. However, the lag in destabilization does not last long, and destabilization soon dominates. To illustrate these principles, we simulated the repression time course of a rapidly induced miRNA for which 80% of the steady-state repression is through mRNA destabilization and 20% is through translational repression (Figure 4). In our simulation, translational repression begins immediately upon miRNA–mRNA association, and mRNA degradation occurs through an increased degradation rate for the miRNA-bound mRNA. This approach yields an early phase in which translational repression dominates, consistent with that observed in our experimental time courses (Figure 4B). The transition from mostly translational repression to mostly mRNA destabilization takes place at 5.7 h (Figure 4C), when relatively little overall repression (9.7% RPF decrease, compared to a 50% decrease at steady state) is occurring (Figure 4B). Our example simulates very rapid miRNA induction; within 6 h the induced miRNA reaches levels that would make it the highest expressed miRNA in 3T3 cells (Figure 4A), similar to or faster than the induction observed in our 3T3 cell lines (Figure 2A). Slowing the induction rate by about half would result in this transition occurring at a point of even less repression (6.6% RPF decrease), and thus in most mammalian contexts, miRNA induction would be too slow to yield detectable repression during the phase in which TE changes dominate. For an early phase of substantial repression mediated primarily through TE changes, miRNA induction would have to be stronger than that ever reported, which is consistent with our inability to find a mammalian context with substantial translation-based repression.
Figure 4. Simulated Dynamics of miRNA-Mediated Repression.

(A) Simulation of rapid miRNA induction that begins with no miRNA and rises to a concentration exceeding that of the highest expressed endogenous miRNA in 3T3 cells within 6 h.
(B) Changes in target mRNA (blue), TE (green), RPF (black), and protein (red; solid line, 10 h protein half-life; dashed line, 100 h protein half-life) levels resulting from the miRNA induction in panel A.
(C) The relative contributions of mRNA decay and translational repression to the overall repression in panel A, when measured at either the RPF level (dark blue and dark green, respectively) or the protein level (light blue and light green; solid lines, 10 h protein half-life; dashed lines, 1 h protein half-life).
Decreases in mRNA and TE lead to decreased protein from the targeted messages, and this change in protein is what matters to the cell. Despite the ultimate importance of the protein changes, measuring these changes over time is less informative for analyzing miRNA repression dynamics than is measuring RPF and mRNA changes, which more directly captures the molecular effects of the miRNA in inhibiting translation and destabilizing mRNA. RPF and mRNA measurements are also more suitable for quantitative comparisons for two reasons: 1) they enabled accurate comparisons of more miRNA targets, and 2) they were each acquired using analogous methods that measured differences at one moment in time without the complications that arise from pre-steady-state measurements of protein changes. With regard to these complications, protein differences detected using direct labeling or standard metabolic labeling (e.g., SILAC) cannot distinguish between protein synthesized before or after induction of the miRNA and thus are unsuitable for pre-steady-state measurements because they would underestimate the impact on newly synthesized protein. Pulsed SILAC differentiates between pre-existing and newly synthesized protein but as currently implemented still entails an extended period (≥6 h for global measurements) of metabolic labeling (Schwanhausser et al., 2009; Huo et al., 2012), which compromises its utility for observing the results of the first few hours of repression.
Despite the advantages of measuring RPF and mRNA changes, we note that during pre-steady-state conditions, the relative TE and mRNA effects can underestimate the relative contribution of translational repression to miRNA-mediated repression at the protein level. For example, at a given time point reduced mRNA might explain 80% of the RPF effect, leaving only 20% of the reduced protein synthesis at that moment to be explained by translational repression, but when considering the reduced protein levels (not current protein synthesis) more repression might have been due to translational repression. This is because the reduced protein levels are a function of the miRNA effects integrated up to the current time point, which includes earlier periods in which translational repression might have represented a greater share of the decreased protein synthesis.
The extent to which the relative contribution of translational repression would be underestimated depends on three factors: 1) the extent to which translational repression represents a greater share of the overall repression at the earlier time periods, 2) the relative strength of the overall repression during earlier periods, and 3) the stability of the protein. Our results indicate that with respect to the second factor, the relative strength of the overall repression during earlier periods is low in mammalian contexts, which implies that any underestimate of the contribution of translational repression to the reduction in protein levels would be minimal. In our simulation the greatest underestimate was observed at 5.7 h, when TE changes explained 49% of the reduction in protein synthesis at that moment and 58% of the reduction in protein accumulation, assuming intermediate protein stability (10 h protein half-life; Figure 4C). A shorter protein half-life further diminished the small differential between protein synthesis and protein accumulation (Figure 4C), whereas a longer half-life delayed the onset of any consequential miRNA effect on protein abundance to a period well beyond the onset of substantial mRNA decay (Figure 4B). In sum, monitoring protein levels rather than TE would not increase the prospects for finding a mammalian setting in which substantial translational repression dominates.
Comparison of fish embryos and mammalian contexts
Attempts to characterize the dynamics of the two modes of miRNA-mediated repression in zebrafish embryos were confounded by two unique features of fish and frog embryos prior to gastrulation: 1) a strong coupling between poly(A)-tail length and translational efficiency and 2) an unusual mRNA metabolism wherein mRNAs with short poly(A)-tails are stable. These features do not necessarily preclude analysis of dynamics, but in these contexts changes in TE due to miRNA-mediated deadenylation must be accounted for independently of changes in TE due to direct miRNA-mediated translational repression. Indeed, when the repression due to mRNA decay is thought of as including deadenylation-dependent translational repression, mRNA decay is the predominant mode of miRNA-mediated repression at all time points analyzed in zebrafish (Subtelny et al., 2014) just as it is at all but the earliest time points in mammalian cells. An important difference between most mammalian systems and early developmental systems (and presumably neuronal synapses or other systems with the aforementioned features) is that, in the latter, effects on translation must be measured to accurately capture the impact of the miRNA on gene expression, and effects on deadenylation must be measured to understand how repression is achieved. However, neither system seems to have a phase in which deadenylation-independent translational repression performs substantial repression without even stronger repression detectable by mRNA changes.
Mechanistic interpretations
Although translational repression and mRNA decay both lead to reduced protein synthesis, the mechanism used for repression has important biological implications. To the extent that repression occurs through translational repression, rapid recovery would be possible without requiring new transcription. This would, for example, be the case in early zebrafish embryos, where the repression of miRNA targets could be rapidly reversed through cytoplasmic polyadenylation. In most settings, however, reversal of miRNA-mediated repression requires new transcription, as mRNA decay constitutes the major mode of repression.
When miRNA-mediated mRNA decay was first reported, it was proposed to occur either through active recruitment of mRNA-degradation machinery or as a secondary effect of inhibiting translation (Lim et al., 2005). Although we observe translational repression prior to the decay of endogenous mRNAs in some experiments, this temporal relationship does not imply that mRNA decay is a consequence of translational repression, because it is also consistent with mRNA decay simply being a slower process. Indeed, several observations favor the model that the decay occurs through active recruitment of mRNA-degradation machinery rather than as a secondary effect of inhibiting translation. First, miRNA targeting can destabilize reporter transcripts that cannot be translated, which indicates that mRNA destabilization is not merely a secondary effect of reducing the number of ribosomes translating an mRNA (Mishima et al., 2006; Wu et al., 2006; Eulalio et al., 2007; Wakiyama et al., 2007; Eulalio et al., 2009; Fabian et al., 2009), although it does not rule out models in which only translationally repressed mRNAs can be destabilized. Second, direct biochemical interactions link miRNAs to Argonaute, Argonaute to TNRC6, and TNRC6 to the deadenylase complexes (the PAN2-PAN3 complex and the CCR4-NOT complex) that shorten the poly(A) tail (Braun et al., 2012), thereby showing how the mRNA-degradation machinery can be actively recruited independent of either the act or the consequence of translational repression. Finally, our work greatly expands the number of mammalian systems examined and shows that in each of these systems mRNA destabilization explains a large majority (from 66% to >90%) of the miRNA-mediated repression observed at steady state.
The idea that the mRNA destabilization might be a secondary consequence of inhibiting translation would be more plausible if a larger fraction of the steady-state repression was through translational repression; otherwise, the mRNA destabilization is out of proportion to the translational repression. We are not aware of any mammalian examples in which translationally repressed messages are so destabilized as a secondary consequence of this repression that the amount of steady-state destabilization exceeds the amount of steady-state translational repression. Indeed, the idea that mammalian messages might be destabilized solely as a secondary consequence of reduced ribosome occupancy or density appears to be largely an extrapolation from observations made in bacteria and yeast, but not mammalian cells (Muhlrad et al., 1995; Schwartz and Parker, 1999; Deana and Belasco, 2005). When examining mammalian mRNAs in general (irrespective of miRNA targeting), we find only a very weak correlation between TE and mRNA half-life (Figure S4B, R2 = 0.004 and 0.001 for 3T3 and HeLa, respectively), and others have shown that repression of translational initiation through the iron response element (a textbook example of mammalian translational repression) does not impart detectable destabilization of either its endogenous host mRNAs (Coccia et al., 1992; Melefors et al., 1993; Kim et al., 1996) or a reporter transcript (Hentze et al., 1987). Thus, when considered together, the available evidence strongly supports a model in which miRNAs actively recruit the deadenylation machinery, and the ensuing deadenylation, decapping, and decay comprises the major mode of miRNA-mediated repression of endogenous targets in mammalian cells.
Some translational repression accompanies mRNA destabilization as a minor component of endogenous target repression in mammalian cells. Like mRNA destabilization, this translational repression also appears to depend on recruitment of CCR4-NOT, but three observations indicate that this repression is not simply a consequence of shortened poly(A)-tails. First, mRNAs without poly(A) tails can be translationally repressed (Wu et al., 2006; Eulalio et al., 2008; Eulalio et al., 2009; Braun et al., 2011; Chekulaeva et al., 2011; Zekri et al., 2013). Second, mutant complexes lacking deadenylase activity can nonetheless promote translational repression (Cooke et al., 2010). Third, tail length and TE are not correlated in most mammalian settings (Subtelny et al., 2014). Thus, the two modes of miRNA-mediated repression seem to represent two independent ramifications of recruiting the deadenylation complexes.
Reconciling results with single-gene studies of mRNA and protein changes
The conclusion that mRNA destabilization is the major mode of miRNA-mediated repression agrees with many previous observations monitoring protein and mRNA changes of single target genes after perturbing a miRNA. Among the >30,000 research studies of mammalian miRNAs, there are also counter examples, in which single-gene measurements seem to suggest a greater role for translational repression (Poy et al., 2004; O'Donnell et al., 2005; Zhao et al., 2005; Chen et al., 2006). An advantage of our approach is that we simultaneously examine thousands of genes, comparing the changes of both mRNA level and TE for hundreds of genes that have at least one miRNA site to those of hundreds of genes that lack a site and thus serve as internal controls. The aggregate result of this global approach should reflect the overall contributions of mRNA destabilization and translational repression, whereas a single-gene study might choose a non-representative example and reach a conclusion that does not apply more generally to the targets of the miRNA.
This raises the question as to what might explain a single-gene result in which a miRNA-dependent change is observed in protein (i.e., with an immunoblot) but not mRNA (e.g., with quantitative reverse-transcription–PCR), which would appear as an outlier in our analyses. Might such outliers represent targets that are repressed at the level of translation without being destabilized? Although this possibility cannot be excluded, changes observed among our control genes that lack miRNA sites raise doubts about its validity. In most experiments (the possible exception being U2OS cells transfected with miR-155), a similar number of these control genes also change at the level of translation without being destabilized (Figure 1D). The observation that this behavior usually does not depend on the presence of a site to the miRNA suggests that either indirect effects of the miRNA or experimental variability explain the presence of most outliers that appear to be changing only at the level of translation.
Other single-gene examples for which translational repression is reported to be the major mode of miRNA-mediated regulation examine reporter mRNAs rather than endogenous mRNAs (Doench and Sharp, 2004; Kiriakidou et al., 2004; Nelson et al., 2004; Yekta et al., 2004; Pillai et al., 2005). Interestingly, the fractional component of regulation attributable to translational repression generally seems to be higher for reporters than for endogenous genes. We have begun experiments that aim to understand this difference between reporter and endogenous genes. Once this difference is understood, reporters could be developed that better recapitulate the regulation of endogenous genes, which would provide more relevant tools for studying the mechanism and dynamics of miRNA-mediated repression.
Experimental Procedures
RNA Isolation
For RNA-seq, total RNA was extracted from B cells and U2OS cells using TRI reagent. Using TRI reagent, cytoplasmic RNA was extracted from cytoplasmic fractions of U2OS cells that were separated from nuclear fractions by differential centrifugation. Briefly, whole-cell lysate prepared as described (Guo et al., 2010) was centrifuged at 1,300g for 10 min, and the resulting supernatant was collected as the cytoplasmic fraction while the pellet obtained was collected as the nuclear fraction. To prepare rRNA/tRNA-depleted U2OS total RNA, total RNA was first treated with the Ribo-Zero rRNA removal kit (Epicentre BioTechnologies) according to manufacturer’s instructions. The resulting rRNA-depleted RNA sample was then spin-filtered using Ultra-4 centrifugal filters with Ultracel-100 membranes (Amicon) by centrifuging at 5,000g for 10 min at 4°C. The filtrate was enriched in tRNAs and was discarded, and the retentate was collected as the rRNA/tRNA-depleted RNA sample. RNA for all other RNA-seq samples was prepared by extracting RNA from ribosome profiling lysates with TRI reagent as described (Subtelny et al., 2014). Except in the case of the tRNA/rRNA-depleted U2OS RNA sample, the extracted RNA was poly(A)-selected as described (Subtelny et al., 2014).
Ribosome Footprint Profiling and RNA-seq
For B cell and U2OS samples, ribosome profiling and RNA-seq were performed essentially as described (Guo et al., 2010), with the only difference being how the RNA was isolated or enriched the cases of U2OS cytoplasmically enriched RNA and tRNA/rRNA-depleted total RNA. All other samples were prepared as described (Subtelny et al., 2014). Detailed protocols are available at http://bartellab.wi.mit.edu/protocols.html. Reference transcript annotations were downloaded (in refFlat format) from the UCSC Genome browser, and for each gene, the longest transcript was chosen as a representative transcript model. RPF and RNA-seq reads were mapped to ORFs as described, which excluded the first 50 nt of each ORF so as to eliminate signal from ribosomes that initiated after adding cycloheximide (Subtelny et al., 2014).
Accession numbers
The NCBI GEO accession number for all microarray and sRNA-seq data and most ribosome profiling and RNA-seq data is GSE61073. The accession number for HeLa and miR-223 neutrophil data analyzed in this study is GSE22004. The accession number for the U2OS ribosome profiling data and RNA-seq data from poly(A)-selected total RNA and tRNA/rRNA-depleted total RNA is GSE51584. The accession numbers for HEK293T mock-treated RNA-seq and ribosome profiling data are GSM1276541 and GSM1276542, respectively. The accession numbers for the uninduced miR-155 actively dividing 3T3 RNA-seq and ribosome profiling data are GSM1276543 and GSM1276544, respectively.
Supplementary Material
Highlights.
In diverse cell types and conditions, mRNA decay is the dominant miRNA effect
Just after miRNA induction, translational repression contributes more of the repression
However, by the time substantial repression is observed, decay is the dominant effect
A mathematical model provides a framework for miRNA-mediated repression
Acknowledgements
We thank V. Agarwal and V. Auyeung for helpful discussions, D. Patrick, E. van Rooji and E. Olson for miR-21 knockout mice and wild-type controls, and the Whitehead Genome Technology Core for sequencing. Supported by NIH grants R01GM067031 (D.P.B.) and R01CA193244 (K.G.). H.G. was supported by the Agency for Science, Technology and Research, Singapore. D.P.B. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzini AA, Lee MT, Giraldez AJ. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science. 2012;336:233–237. doi: 10.1126/science.1215704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethune J, Artus-Revel CG, Filipowicz W. Kinetic analysis reveals successive steps leading to miRNA-mediated silencing in mammalian cells. EMBO Rep. 2012;13:716–723. doi: 10.1038/embor.2012.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun JE, Huntzinger E, Fauser M, Izaurralde E. GW182 proteins directly recruit cytoplasmic deadenylase complexes to miRNA targets. Mol Cell. 2011;44:120–133. doi: 10.1016/j.molcel.2011.09.007. [DOI] [PubMed] [Google Scholar]
- Braun JE, Huntzinger E, Izaurralde E. A molecular link between miRISCs and deadenylases provides new insight into the mechanism of gene silencing by microRNAs. Cold Spring Harb Perspect Biol. 2012;4 doi: 10.1101/cshperspect.a012328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castilla-Llorente V, Spraggon L, Okamura M, Naseeruddin S, Adamow M, Qamar S, Liu J. Mammalian GW220/TNGW1 is essential for the formation of GW/P bodies containing miRISC. J Cell Biol. 2012;198:529–544. doi: 10.1083/jcb.201201153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekulaeva M, Mathys H, Zipprich JT, Attig J, Colic M, Parker R, Filipowicz W. miRNA repression involves GW182-mediated recruitment of CCR4-NOT through conserved W-containing motifs. Nat Struct Mol Biol. 2011;18:1218–1226. doi: 10.1038/nsmb.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Zheng D, Xia Z, Shyu AB. Ago-TNRC6 triggers microRNA-mediated decay by promoting two deadenylation steps. Nat Struct Mol Biol. 2009;16:1160–1166. doi: 10.1038/nsmb.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coccia EM, Profita V, Fiorucci G, Romeo G, Affabris E, Testa U, Hentze MW, Battistini A. Modulation of ferritin H-chain expression in Friend erythroleukemia cells: transcriptional and translational regulation by hemin. Mol Cell Biol. 1992;12:3015–3022. doi: 10.1128/mcb.12.7.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke A, Prigge A, Wickens M. Translational repression by deadenylases. J Biol Chem. 2010;285:28506–28513. doi: 10.1074/jbc.M110.150763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deana A, Belasco JG. Lost in translation: the influence of ribosomes on bacterial mRNA decay. Genes Dev. 2005;19:2526–2533. doi: 10.1101/gad.1348805. [DOI] [PubMed] [Google Scholar]
- Djuranovic S, Nahvi A, Green R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science. 2012;336:237–240. doi: 10.1126/science.1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Izaurralde E. GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat Struct Mol Biol. 2008;15:346–353. doi: 10.1038/nsmb.1405. [DOI] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Nishihara T, Rehwinkel J, Fauser M, Izaurralde E. Deadenylation is a widespread effect of miRNA regulation. RNA. 2009;15:21–32. doi: 10.1261/rna.1399509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Rehwinkel J, Stricker M, Huntzinger E, Yang SF, Doerks T, Dorner S, Bork P, Boutros M, Izaurralde E. Target-specific requirements for enhancers of decapping in miRNA-mediated gene silencing. Genes Dev. 2007;21:2558–2570. doi: 10.1101/gad.443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR, Mathonnet G, Sundermeier T, Mathys H, Zipprich JT, Svitkin YV, Rivas F, Jinek M, Wohlschlegel J, Doudna JA, et al. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol Cell. 2009;35:868–880. doi: 10.1016/j.molcel.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia DM, Baek D, Shin C, Bell GW, Grimson A, Bartel DP. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat Struct Mol Biol. 2011;18:1139–1146. doi: 10.1038/nsmb.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, Hammond SM, Bartel DP, Schier AF. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson DG, Hogan DJ, McCullough HL, Myers JW, Herschlag D, Ferrell JE, Brown PO. Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS Biol. 2009;7:e1000238. doi: 10.1371/journal.pbio.1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze MW, Rouault TA, Caughman SW, Dancis A, Harford JB, Klausner RD. A cis-acting element is necessary and sufficient for translational regulation of human ferritin expression in response to iron. Proc Natl Acad Sci U S A. 1987;84:6730–6734. doi: 10.1073/pnas.84.19.6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtz J, Pasquinelli AE. Uncoupling of lin-14 mRNA and protein repression by nutrient deprivation in Caenorhabditis elegans. RNA. 2009;15:400–405. doi: 10.1261/rna.1258309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo Y, Iadevaia V, Yao Z, Kelly I, Cosulich S, Guichard S, Foster LJ, Proud CG. Stable isotope-labelling analysis of the impact of inhibition of the mammalian target of rapamycin on protein synthesis. Biochem J. 2012;444:141–151. doi: 10.1042/BJ20112107. [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HY, LaVaute T, Iwai K, Klausner RD, Rouault TA. Identification of a conserved and functional iron-responsive element in the 5'-untranslated region of mammalian mitochondrial aconitase. J Biol Chem. 1996;271:24226–24230. doi: 10.1074/jbc.271.39.24226. [DOI] [PubMed] [Google Scholar]
- Kiriakidou M, Nelson PT, Kouranov A, Fitziev P, Bouyioukos C, Mourelatos Z, Hatzigeorgiou A. A combined computational-experimental approach predicts human microRNA targets. Genes Dev. 2004;18:1165–1178. doi: 10.1101/gad.1184704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with 'antagomirs'. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Melefors O, Goossen B, Johansson HE, Stripecke R, Gray NK, Hentze MW. Translational control of 5-aminolevulinate synthase mRNA by iron-responsive elements in erythroid cells. J Biol Chem. 1993;268:5974–5978. [PubMed] [Google Scholar]
- Mishima Y, Giraldez AJ, Takeda Y, Fujiwara T, Sakamoto H, Schier AF, Inoue K. Differential regulation of germline mRNAs in soma and germ cells by zebrafish miR-430. Curr Biol. 2006;16:2135–2142. doi: 10.1016/j.cub.2006.08.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D, Decker CJ, Parker R. Turnover mechanisms of the stable yeast PGK1 mRNA. Mol Cell Biol. 1995;15:2145–2156. doi: 10.1128/mcb.15.4.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson PT, Hatzigeorgiou AG, Mourelatos Z. miRNP:mRNA association in polyribosomes in a human neuronal cell line. RNA. 2004;10:387–394. doi: 10.1261/rna.5181104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- Olsen PH, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev Biol. 1999;216:671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- Rehwinkel J, Behm-Ansmant I, Gatfield D, Izaurralde E. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA. 2005;11:1640–1647. doi: 10.1261/rna.2191905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rissland OS, Hong SJ, Bartel DP. MicroRNA destabilization enables dynamic regulation of the miR-16 family in response to cell-cycle changes. Mol Cell. 2011;43:993–1004. doi: 10.1016/j.molcel.2011.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhausser B, Gossen M, Dittmar G, Selbach M. Global analysis of cellular protein translation by pulsed SILAC. Proteomics. 2009;9:205–209. doi: 10.1002/pmic.200800275. [DOI] [PubMed] [Google Scholar]
- Schwartz DC, Parker R. Mutations in translation initiation factors lead to increased rates of deadenylation and decapping of mRNAs in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:5247–5256. doi: 10.1128/mcb.19.8.5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seggerson K, Tang L, Moss EG. Two genetic circuits repress the Caenorhabditis elegans heterochronic gene lin-28 after translation initiation. Dev Biol. 2002;243:215–225. doi: 10.1006/dbio.2001.0563. [DOI] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Subtelny AO, Eichhorn SW, Chen GR, Sive H, Bartel DP. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature. 2014;508:66–71. doi: 10.1038/nature13007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- Wakiyama M, Takimoto K, Ohara O, Yokoyama S. Let-7 microRNA-mediated mRNA deadenylation and translational repression in a mammalian cell-free system. Genes Dev. 2007;21:1857–1862. doi: 10.1101/gad.1566707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci U S A. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- Zekri L, Kuzuoglu-Ozturk D, Izaurralde E. GW182 proteins cause PABP dissociation from silenced miRNA targets in the absence of deadenylation. Embo J. 2013;32:1052–1065. doi: 10.1038/emboj.2013.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.