Abstract
Higher eukaryote genomes contain repetitive DNAs, often concentrated in transcriptionally inactive heterochromatin. Although repetitive DNAs are not typically considered as regulatory elements that directly affect transcription, they can contain binding sites for some transcription factors. Here, we demonstrate that binding of the transcription factor CCAAT/Enhancer Binding Protein alpha (C/EBPα) to the mouse major α-satellite repetitive DNA sequesters C/EBPα in the transcriptionally inert pericentromeric heterochromatin. We find that this sequestration reduces the transcriptional capacity of C/EBPα. Functional sequestration of C/EBPα was demonstrated by experimentally reducing C/EBPα binding to the major α-satellite DNA, which elevated the concentration of C/EBPα in the non-heterochromatic subcompartment of the cell nucleus. The reduction in C/EBPα binding to α-satellite DNA was induced by the co-expression of the transcription factor Pit-1, which removes C/EBPα from the heterochromatic compartment, and by the introduction of an altered-specificity mutation into C/EBPα that reduces binding to α-satellite DNA but permits normal binding to sites in some gene promoters. In both cases, the loss of α-satellite DNA binding coincided with an elevation in the binding of C/EBPα to a promoter and an increased transcriptional output from that promoter. Thus, the binding of C/EBPα to this highly repetitive DNA reduced the amount of C/EBPα available for binding to and regulation of this promoter. The functional sequestration of some transcription factors through binding to repetitive DNAs may represent an underappreciated mechanism controlling transcription output.
The eukaryotic cell nucleus is highly structured into subdomains of specialized function that include compartments of high and low transcriptional capacity, the euchromatin and heterochromatin (reviewed in (1–4). The most transcriptionally inactive compartment, constitutive heterochromatin, consists typically of repetitive DNAs and specialized, highly compact chromatin concentrated near centromeres or telomeres (5, 6). Although repetitive DNAs retain their historic label as ‘junk’ DNA, they play an important role in chromosomal segregation as an essential structural component of the centromere (7). In addition, the heterochromatic chromosomal domains repress the activity of nearby genes (2, 8). This indicates an indirect role for repetitive DNA in transcriptional regulation but it remains unknown if repetitive DNAs can somehow directly affect transcription.
A number of transcription factors concentrate at heterochromatic regions (9–18). Where examined, the concentration of transcription factors at heterochromatin depends upon retention of their DNA binding domain (14, 16, 19–21). This suggests that direct DNA binding, possibly to the repetitive DNAs, may target those transcription factors to the heterochromatin. Indeed, some transcription factors can bind specific repetitive elements in vitro (11, 16–19). The correlation between repetitive DNA binding and targeting to heterochromatin is strengthened by a report that treatment of a preadipocyte cell line with growth hormone enhances targeting of the transcription factor CCAAT/Enhancer Binding Protein beta (C/EBPβ2 to heterochromatin and the ability of that factor to bind a repetitive DNA in vitro (17). Furthermore, Drosophila embryos undergo homeotic transformations when treated with polyamide drugs that bind a repetitive DNA element to which the GAGA factor binds (20, 22). However, establishing if there is a causal or coincidental relationship between repetitive DNA binding, heterochromatic targeting and biologic response of a site-specific transcription factor has been difficult to address experimentally (1, 3, 20, 22–24). In the current study, we identify methods to preferentially block the binding of a transcription factor to repetitive DNA, and then follow the effects of that intervention on promoter binding and activation.
CCAAT/enhancer binding protein alpha (C/EBPα) is a transcription factor important to the differentiation and transcriptional regulation of a number of cell types (25–28). In addition to binding and activating a large number of gene promoters, C/EBPα also is capable of binding in vitro to the mouse major α-satellite repetitive DNA (11). The major α-satellite DNA repeat (referred to hereafter as α-satellite DNA) is prevalent at murine pericentromeric heterochromatin (29, 30). Endogenous C/EBPα expressed upon differentiation of mouse 3T3-L1 cells into adipocytes concentrates strongly at the pericentromeric heterochromatin (11, 31), possibly through its binding to α-satellite DNA. The concentration of C/EBPα at pericentromeric heterochromatin is an intrinsic property of C/EBPα as C/EBPα ectopically expressed in undifferentiated 3T3-L1 pre-adipocyte cells and in pituitary progenitor cells also concentrates there (31, 32). The ability to mimic the intranuclear localization with ectopically expressed proteins enabled us to manipulate C/EBPα and establish that the conserved bZIP domain of C/EBPα was necessary and sufficient for ectopically expressed C/EBPα to target to pericentromeric heterochromatin (21, 33). We also showed previously that the concentration of C/EBPα in transcriptionally inactive heterochromatin is highly regulated by the pituitary-specific transcription factor Pit-1 which translocates C/EBPα from a pericentromeric to a non-pericentromeric distribution in the pituitary progenitor cell model (31, 34, 35).
Here, we provide experimental evidence that specific binding of C/EBPα binding to α-satellite DNA sequesters C/EBPα in the pericentromeric heterochromatin of the cell nucleus. Furthermore, the regulated translocation of C/EBPα away from the pericentromeric heterochromatin (31, 34, 35) correlated with a loss of C/EBPα binding to α-satellite DNA, enhanced promoter binding and enhanced promoter activation. This indicates that transcription factor binding to repetitive DNA can act as a direct negative regulator of transcriptional activity. The impact of repetitive DNA binding on the activity of other transcription factors remains unknown but this report suggests that it is a facet of transcription regulation that should be considered for some factors.
EXPERIMENTAL PROCEDURES
Imaging
Cells were stained with anti-C/EBPα ‘C-18’ (Santa Cruz Biotechnology sc-9314, Santa Cruz, CA) and a rhodamine-linked anti-IgG secondary antibody, then counterstained with Hoechst 33342 (Invitrogen, Carlsbad, CA) as described (32). Similar results were obtained with anti-C/EBPα antibodies sc-9315 or sc-61 (Santa Cruz Biotechnology). Representative images were acquired confocally with a 63x/1.40 NA oil immersion objective (Leica, Wetzlar, Germany)(Fig. 1). To quantify the location of C/EBPα relative to pericentromeric heterochromatin (Fig. 2B), widefield microscope images were acquired using a 40x/0.95 NA air objective (Olympus, Tokyo, Japan). The relative positions of the anti-C/EBPα-stained material and the Hoechst 33342-stained pericentromeric heterochromatin were quantified from >50 cells for each mutant using the correlation plot functions of Metamorph (Molecular Devices Corporation, Downingtown, PA) as described (32). For these quantitative studies, cells were stained with the M2 antibody against the FLAG epitope (Sigma, St. Louis, MO) that was added to the amino terminus of all mutants to ensure equivalent immuno-detection of the different mutants.
FIGURE 1.
C/EBPα concentrates at pericentromeric heterochromatin. A, B, Wild-type and C, D, mutant C/EBPα were transiently expressed in mouse 3T3-L1 cells (A, C) or pituitary progenitor GHFT1-5 cells (B, D) at levels similar to that found following differentiation of the 3T3-L1 cells to adipocytes. Wild-type C/EBPα, detected by anti-C/EBPα antibody staining, concentrates in defined subregions of the cell nucleus that coincide with pericentromeric heterochromatin stained by the blue fluorescent DNA binding dye Hoechst 33342. C/EBPα containing valine 296 mutated to alanine (V296A) does not concentrate at pericentromeric heterochromatin. The cells and images in this figure were stained in parallel and captured/displayed under identical size and image processing conditions.
FIGURE 2.
Mutations that disrupt C/EBPα concentration at pericentromeric heterochromatin also disrupt binding to α-satellite DNA. A, Diagram of the primary structure of C/EBPα and the position of point mutations used in this study relative to landmarks. TA domains, transcriptional activation domains; b, basic region; ZIP, leucine zipper. Blocks of amino acids conserved amongst C/EBP proteins from different species and with selected bZIP proteins are indicated as a black bar. Numbers represent amino acid positions. C/EBPα mutants are named according to the amino acids that were mutated (e.g. X60-62). The known activities affected by these mutations are briefly described in the Results section. B, Co-localization of antibody-stained C/EBPα with Hoechst-33342 stained C/EBPα was quantified from multiple cells as a correlation coefficient for wild-type (WT) and mutant C/EBPα expressed in GHFT1-5 cells (mean +/− sd, n>50 cells for each). A correlation coefficient of +1.0 indicates complete co-localization; 0.0 indicates no relationship; −1.0 indicates completely opposite localization. *, mutants with correlation coefficients significantly (p<<0.001) less than for wild-type C/EBPα. There is no correlation coefficient for cells transfected with vector alone since there is no C/EBPα present. C, Expression level of the same mutants monitored by western blots of nuclear extracts from transfected GHFT1-5 cells (mean quantified by densitometry from three independent experiments). D, Representative gel mobility shift of radiolabelled α-satellite DNA by wild-type C/EBPα and the indicated mutants present in the nuclear extracts of GHFT1-5 cells. The complexes formed with wild-type C/EBPα were supershifted by an antibody against C/EBPα (not shown).
Ectopic Expression of C/EBPα
GHFT1-5 cells and 3T3-L1 cells were grown as previously described (31). GHFT1-5 cells were transfected with the Rc-C/EBPα expression vector (FLAG-epitope-tagged C/EBPα cDNA subcloned into the Rc/CMV expression vector of Invitrogen) using the Fugene6 reagent (Roche Applied Science, Basel, Switzerland). 3T3-L1 cells were transfected using the Lipofectamine Plus reagent (Invitrogen). Point mutations were introduced into the C/EBPα expression vector using QuickChange (Stratagene, La Jolla, CA) and were confirmed by complete sequencing of the C/EBPα cDNA. The C/EBPα-Gal expression vectors were constructed by inserting the Gal4 DNA binding domain (amino acids 1–147) amplified from the pM vector (Clontech, Palo Alto, CA) in frame between C/EBPα amino acids 308 and 309 in the Rc-C/EBPα vector. The nucleotide sequence of the three amino acid long spacer linking C/EBPα amino acid 308 (underlined) to Gal4 amino acid 1 (underlined) is CTCAAGCTTCCCAAG.
Extract Preparation and Western Blotting
Cytoplasmic and nuclear extracts were prepared from cells transfected with the Rc/CMV, Rc-C/EBPα wild-type or mutant expression vectors, as previously described (36). Protein amount in the extracts was determined by the Coomassie Plus protein assay (Pierce Chemical Company, Rockford, IL). Expression of C/EBPα and mutants was assessed by western blots of equal amounts of protein in each extract using the anti-FLAG M2 antibody (Sigma) or the anti-C/EBPα 14AA antibody (Santa Cruz Biotechnology sc-61), a species-appropriate horseradish peroxidase-linked anti-IgG secondary antibody (Santa Cruz Biotechnology) and the enhanced chemiluminescence reagent (Amersham Biosciences, Amersham, U.K.). Levels of expression for the different mutants (Fig. 2C) in the nuclear extracts were quantified by densitometry of unsaturated films from anti-FLAG-probed extracts from three independent experiments. None of the C/EBPα mutants were observed in the cytoplasmic extracts.
DNA Binding
Nuclear extracts were used in gel mobility shift assays of radiolabelled α-satellite DNA (Figs. 2D, 3C) or of radiolabelled oligonucleotides representing the previously described C/EBPα binding sites found at −230 in the growth hormone promoter (37) or at −68 in the PU.1 promoter (38)(Figs. 3A, 3B, 5B). α-satellite DNA was prepared by polymerase chain reaction from total genomic DNA extracted from GHFT1-5 cells using the primers TGGAATATGGCGAGATAACTGAAAATCACGG and AGGTCCTTCAGTGTGCATTTCTCATTT. The 234 bp single copy of α-satellite DNA was gel purified from higher multiples of this tandem repetitive element and represents the population of α-satellite DNAs present in those cells. Similar results were obtained when the gel shift experiments were conducted with subcloned, single-copy, radiolabelled α-satellite DNAs (not shown). Specific C/EBPα binding to all sites was demonstrated by supershifts of the gel-shifted bands with the anti-FLAG M2 or 14AA antibodies, and by competition with excess unlabeled α-satellite DNA (not shown). Quantitative comparison of subsaturating equilibrium binding by wild-type and V296A (Fig. 3) was done by mixing 4 µg total extract, 2 µg poly dI-dC and radiolabelled DNA. The amounts of probe bound by wild-type and mutant C/EBPα were quantified by densitometry of unsaturated films using extracts of three independent transfections. The removal of rabbit reticulocyte lysate-translated C/EBPα by the addition of rabbit reticulocyte-translated Pit-1 (Fig. 3C) was done under the same binding conditions.
FIGURE 3.
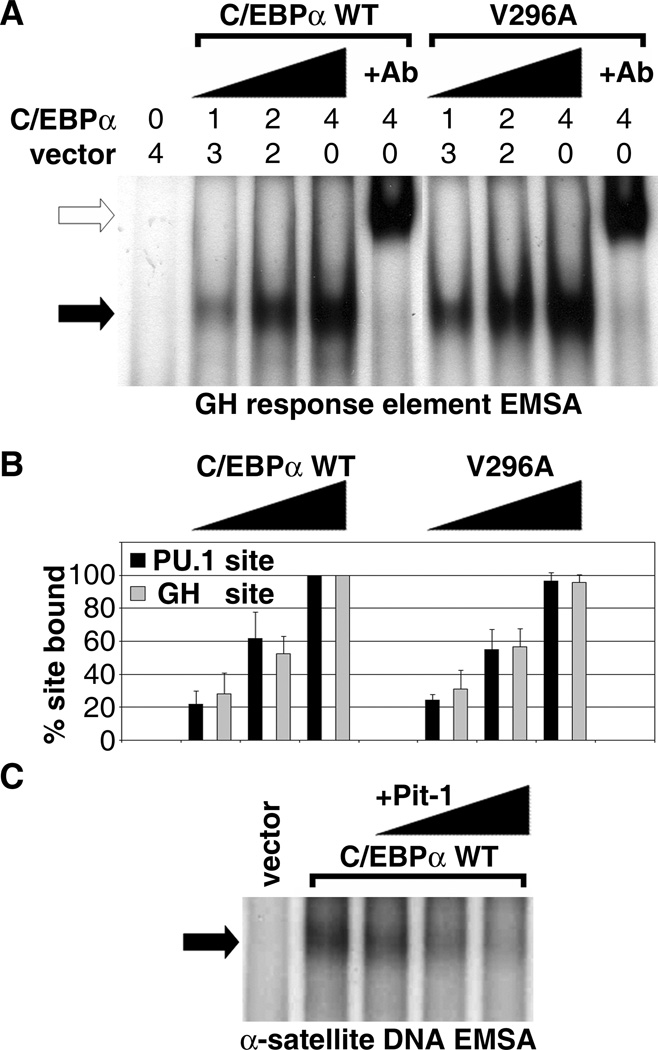
Correlation of pericentromeric targeting with specific binding to α-satellite DNA. A, The V296A mutation that disrupts α-satellite DNA binding does not affect binding to some other response elements. Representative electrophoretic mobility shift (black arrow) of a radiolabelled C/EBP response element from the growth hormone (GH) promoter complexed with subsaturating amounts of extracts (0.0, 1.0, 2.0 and 4.0 µg) of transfected GHFT1-5 cells expressing equivalent levels of wild-type and V296A C/EBPα. Total extract amount was held constant with the addition of 4.0, 3.0, 2.0 or 0.0 µg of extract from sham-transfected cells. The complexes detected were supershifted with an antibody against C/EBPα (open arrow). The wild-type and V296A gel shifts shown are from the same X-ray film with an intervening series of lanes digitally removed. B, Quantitative summary of electrophoretic mobility shift assays evaluating wild-type and V296A mutant binding to the GH and PU.1 response elements (mean +/− sd of three independent experiments). No statistically significant difference (p>0.10 for all concentrations) was observed in the equilibrium binding of wild-type and V296A mutant C/EBPα to either response element. C, The addition of Pit-1, which also inhibits C/EBPα localization to pericentromeric heterochromatin (35), also blocks in vitro binding of C/EBPα to α-satellite DNA. Binding of rabbit reticulocyte-expressed C/EBPα to α-satellite DNA was inhibited in a dose dependent fashion by the addition of reticulocyte-expressed Pit-1. Total reticulocyte lysate amount was held constant by the addition of sham-translated extracts.
FIGURE 5.
Superactivity by the V296A mutation is not associated with enhanced intrinsic transcriptional activity. A, Full-length V296A superactivates the C/EBP-RE reporter (see Fig. 4) in both undifferentiated 3T3-L1 cells and GHFT1-5 cells. *, promoter activation of V296A C/EBPα that is significantly greater than WT C/EBPα. B, Intrinsic transcriptional activity was measured as the amount of luciferase expressed from a promoter containing Gal4 binding sites upon co-expression of fusions with the Gal4 DNA binding domain with amino acids 1–308 of the wild-type or V296A mutant C/EBPα. There was no activation by the Gal4 DBD alone or by either wild-type or mutant C/EBPα (amino acids 1–308). Data represent the mean +/− sd from six, (A, GHFT1-5), four (A, 3T3-L1) or three (B, GHFT1-5 and 3T3-L1) independent experiments. Representative western blots show equivalent amount of expression of the wild-type and V296A mutant C/EBPα proteins.
Reporter Analysis
The cytoplasmic extracts were used for reporter activation studies. The C/EBP-RE reporter (Figs. 4, 5) consists of a single C/EBPα binding site of the rat growth hormone promoter (−239/−219) inserted upstream of the TATA box of the GH promoter (−33/+8) appended to the chloramphenicol acetyltransferase reporter (37). The Gal-RE reporter (Fig. 4) is the modified ‘GK1’ reporter (39), which consists of five copies of the Gal4 binding site inserted upstream of the minimal adenovirus E1b promoter appended to the luciferase reporter. The reporters were co-transfected with the Rc/CMV (‘vector’), Rc-C/EBPα wild-type or mutant expression vectors. The −85/+158 fragment of the PU.1 promoter was inserted in front of the luciferase reporter (38) and co-transfected with the C/EBPα expression vectors and with a FLAG-tagged Pit-1 cDNA in the Rc/CMV expression vector or with an equivalent amount of Rc/CMV itself. The GK1 and PU.1 reporter vectors were kindly provided by Drs. Paul Webb (UCSF, San Francisco, CA) and Alan Friedman (Johns Hopkins University, Baltimore, MD), respectively.
FIGURE 4.
The euchromatically-distributed V296A mutant superactivates a promoter to which V296A and WT C/EBPα bind equivalently. A, Effect of WT or mutant C/EBPα expression in GHFT1-5 cells on chloramphenicol acetyltransferase activity expressed under the control of a single growth hormone C/EBP response element (C/EBP-RE) appended to a TATA box. Superactivation of the C/EBP-RE promoter/reporter by V296A is not detected with the V314P/L315P, L324P and L331P C/EBPα control mutants that, like V296A, are not sequestered in pericentromeric heterochromatin. Data represent the mean +/− sd from four independent experiments. B, Electrophoretic mobility shift assays show that, unlike the V296A mutant, the control mutants are not capable of binding the GH response element in the C/EBP-RE reporter. Additional lanes between the WT and V296A lanes were digitally deleted for this presentation.
Chromatin Immunoprecipitation
Two days following transfection with expression vectors, chromosomal DNA was formaldehyde cross-linked to protein in approximately 10 × 106 intact GHFT1-5 cells for 10 minutes then glycine quenched. Genomic DNA was isolated and sheered to an average length of 200–1000 bp by sonication. 300 µg of sheered DNA was pre-cleared by binding to protein G-Sepharose beads the cross-linked C/EBPα-DNA complexes were precipitated with anti-C/EBPα (C-18) antibodies. The precipitated DNA was heat-released to remove cross-links to the precipitated C/EBPα, and 1/25th of the resulting DNA was amplified by 12 PCR cycles using α-satellite DNA primers (see DNA Binding Analysis for primers) or by 31 PCR cycles using PU.1 primers (CGACCGGAGCAGCAGAAG and CAAGTTCCTGATTTTATCGAAG). The amount of ethidium bromide-stained amplified DNA was measured only for subsaturating rounds of amplification; the 234 bp ‘monomeric’ α-satellite product and the 239 bp PU.1 gene product were quantified against the amounts of DNA amplified from 2.5% of input DNA.
Quantitative RT-PCR
GHFT1-5 cells were transfected with the IRES-GFP vector (Invitrogen) or the same vector containing wild-type or V296A mutant C/EBPα cDNAs. The transfected cells were isolated as green fluorescent cells using a MoFlo cell sorter (DakoCytomation, Fort Collins, CO). RNA was extracted and quantified by real time RT-PCR using 5’ fluorogenic nuclease techniques. RNA expressed from the endogenous PU.1 gene was detected, following initial cDNA synthesis, with amplification primers in exons 3 and 4 (CGTCCTCGATACTCCCATGGT and AAGGTTTGATAAGGGAAGCACATC) and an intron/exon-spanning probe (6FAM-TCAGTCACCAGGTTTCCTACATGCCCC-BHQ1) created by Drs. Scott Kogan and David Ginziger (UCSF Comprehensive Cancer Center). The number of cycles required to reach threshold detection were compared against a similarly detected internal control for HPRT expression (Applied Biosystems Mm00446968_m1, Foster City, CA and similarly designed probes from David Ginziger at the UCSF Comprehensive Cancer Center), and corrected for differing efficiencies of amplification for the PU.1 and HPRT probes determined in cDNA dilution experiments (40).
Statistical Analyses
All values presented in the figures or in the text represent the mean +/− standard deviation from the number of multiple independent experiments indicated in the figure legends. Significantly different p-values presented in the text were determined using paired (Figs. 3–7) or unpaired (Fig. 2) Student’s t-tests.
FIGURE 7.
Pit-1 co-expression reduces C/EBPα binding to α-satellite DNA and increases binding to and activation of the PU.1 promoter. Representative chromatin immunoprecipitation of A, α-satellite and B, PU.1 promoter DNA by anti-C/EBPα antibodies in GHFT1-5 cells transfected with empty vector or vector expressing wild-type C/EBPα, Pit-1 or C/EBPα and Pit-1 together. Input, 2.5% of input DNA. C, Quantification of the amounts of α-satellite DNA or PU.1 promoter precipitated (mean +/− sd from three independent experiments). Inset, representative western blot of nuclear extracts probed with an anti-FLAG antibody that detects both the FLAG-tagged C/EBPα and FLAG-tagged Pit-1. Lane order in the inset is identical to that presented for the ChIP. D, Amounts of luciferase expressed from the −85/+152 PU.1 promoter in extracts of GHFT1-5 cells two days after transfection with empty vector, wild-type or V296A mutant C/EBPα, with or without co-expressed Pit-1 (mean +/− sd from six independent experiments).
RESULTS
Pericentromeric Targeting by C/EBPα depends on DNA binding
We first examined the mechanism by which C/EBPα concentrates at pericentromeric heterochromatin. C/EBPα ectopically expressed in undifferentiated mouse 3T3-L1 pre-adipocyte cells or in mouse GHFT1-5 pituitary precursor cells (at levels similar to those expressed in differentiated 3T3-L1 cells) concentrates in the Hoechst 33342-stained pericentromeric heterochromatin (31, 32)(Figs. 1A, B). Our prior domain deletion studies showed that the conserved, DNA binding bZIP domain of C/EBPα was necessary and sufficient for ectopically expressed C/EBPα to target to pericentromeric heterochromatin in GHFT1-5 (21) and 3T3-L1 cells (33). Those domain deletion studies did not distinguish whether DNA binding, or some other activity in the bZIP domain, was required to target C/EBPα to pericentromeric heterochromatin. We therefore determined the consequences to pericentromeric target of point mutations in functionally significant regions of C/EBPα that are conserved across multiple species and with other C/EBP isoforms and a selection of bZIP family members (Fig. 2A, conserved regions indicated by black bars).
An example of a C/EBPα mutant (V296A) that does not co-localize with pericentromeric heterochromatin in 3T3-L1 or GHFT1-5 pituitary progenitor cells is shown in figures 1C and 1D. This point mutation is located in the basic domain (V296A) that directly binds DNA (41, 42). Point mutations also were introduced into the leucine zipper (V314P/V315P, L324P and L331P) that is required for C/EBPα dimerization and DNA binding (41, 42) and in more amino terminal amino acids required for C/EBPα binding to TFIIB (X67-69A)(43), TBP (X76-78A)(43), to CBP (X63-65A and X146-148A, X.L., F.S and N. Tovchigrechko, unpublished data) or that that weakly affect C/EBPα binding to TFIIB (X60-62A)(43). The extent by which wild-type and each mutant C/EBPα co-localized with pericentromeric heterochromatin in GHFT1-5 cells was quantified rigorously and averaged (+/− sd) from multiple cells as a correlation coefficient (Fig. 2B). A high positive correlation, such as that quantified for wild-type C/EBPα (Fig. 2B, WT, black bar), shows that locations of high amounts of fluorescence emitted from the Hoechst 33342-stained pericentromeric heterochromatin correspond to locations of high fluorescence emitted from antibody-stained C/EBPα. Conversely, a low or negative correlation (for example, Fig. 2B, V296A, gray bar) shows little or inverse correlation between C/EBPα and pericentromeric location within the cell nucleus.
The C/EBPα mutants in the basic region and leucine zipper all failed to colocalize with the pericentromeric heterochromatin (Fig. 2B, *) whereas all of the more amino-terminal mutants remained concentrated at pericentromeric heterochromatin. Therefore, the ability of C/EBPα to concentrate at pericentromeric heterochromatin was selectively reduced by point mutations in the conserved, DNA-binding bZIP domain. The failure of the bZIP mutants to concentrate at pericentromeric heterochromatin was not a consequence of altered expression level since all mutations were similarly expressed as quantified by western blots from extracts collected in three separate experiments (Fig. 2C; see Figs. 6C and 7C for representative western blots).
FIGURE 6.
Reduced binding of the V296A mutant of C/EBPα to α-satellite DNA is accompanied by increased binding to and activation of the PU.1 gene. Representative chromatin immunoprecipitation of A, α-satellite and B, PU.1 promoter DNA by anti-C/EBPα antibodies in GHFT1-5 cells transfected with empty, wild-type or V296A mutant C/EBPα expression vectors. Input, 2.5% of input DNA. C, Quantification of the amounts of α-satellite DNA or PU.1 promoter precipitated (mean +/− sd from four independent experiments). Inset, representative western blot of nuclear extracts probed with an anti-C/EBPα antibody. Lane order in the inset is identical to that presented for the ChIP: vector, C/EBP, V296A. D, Quantitative RT-PCR measurement of PU.1 mRNA amount in GHFT1-5 cells transfected with an IRES-GFP expression vector, or the same vector containing wild-type or mutant C/EBPα inserted upstream of the internal ribosomal entry site. GFP fluorescence was used to sort the transfected cells away from the untransfected cells before RNA collection.
All four C/EBPα point mutants that disrupted concentration at pericentromeric heterochromatin in GHFT1-5 cells also were shown to be defective in DNA binding, as assessed in vitro by electrophoretic mobility shift assays (EMSA) with a radiolabelled α-satellite DNA probe (Fig. 2D). The complexes formed with the wild-type C/EBPα (black arrow) were supershifted with antibodies against C/EBPα and were competed by an excess of unlabeled competitor probe (data not shown). A minor amount of α-satellite DNA binding remained with theV296A mutation. In contrast, the amino terminal C/EBPα point mutants all remained capable of binding to α-satellite DNA. This data showed that the ability to bind DNA correlated with the ability to target to pericentromeric heterochromatin. However, these studies did not examine if specific binding to α-satellite DNA correlated with C/EBPα targeting to pericentromeric heterochromatin.
C/EBPα binding to α-Satellite DNA Specifically Correlates with Pericentromeric Targeting
Certain attributes of the V296A C/EBPα point mutant provided better evidence for a correlation between specific α-satellite DNA binding with pericentromeric targeting. Valine-296 directly contacts DNA in crystallographic structures of a C/EBPα-DNA complex (44) but the homologous amino acid is an alanine in most other DNA-binding members of the bZIP family. Indeed, C/EBPα containing the V296A mutation still binds a consensus C/EBPα binding site (44). Therefore, although defective in binding to α-satellite DNA, the V296A mutant was not generally deficient in DNA binding.
We conducted quantitative EMSA studies which showed that V296A mutant retained wild-type binding to the C/EBP response elements found in the growth hormone gene (37) and the gene for the PU.1 transcription factor (38). A representative EMSA in which sub-saturating levels of equivalent amounts of wild-type C/EBPα and V296A mutant were complexed to the growth hormone (GH) promoter element is shown in figure 3A. The C/EBPα-DNA complexes (black arrow) were supershifted by antibodies against C/EBPα (open arrow) and were competed by an excess of unlabeled probe (not shown). Quantification of binding from three independent studies demonstrated that the V296A and wild-type C/EBPα bound with equal affinity to both the GH and PU.1 elements (Fig. 3B). By contrast, binding of the V296A mutant to the α-satellite DNA (Fig. 2D) and to the C/EBP response elements from the aP2, GLUT4 and PPARγ promoters (data not shown) was disrupted to varying degrees. The failure of the V296A mutant to target pericentromeric heterochromatin therefore may be a consequence of the failure to bind α-satellite DNA or to other, unknown binding sites. The high concentration of α-satellite DNA at the centromere makes it the most likely candidate binding sequence.
Further support for the hypothesis that specific binding to α-satellite DNA targets C/EBPα to the pericentromeric heterochromatin came from complementary studies based upon our prior observations that expression of Pit-1 strongly reduces the concentration of C/EBPα at pericentromeric heterochromatin (34, 35). Co-incubation of in vitro translated C/EBPα with in vitro translated Pit-1 resulted in a dose-dependent loss of C/EBPα binding to α-satellite DNA (Fig. 3C). Similarly, α-satellite DNA binding in gel shifts using extracts of GHFT1-5 pituitary progenitor cells expressing C/EBPα was reduced by co-expression of Pit-1 in those cells (data not shown). Thus, the ability of Pit-1 to inhibit C/EBPα binding to α-satellite DNA, together with the inability of the V296A mutant to bind α-satellite DNA, provided strong complementary evidence that specific C/EBPα binding to α-satellite DNA is the primary event by which C/EBPα localizes to pericentromeric heterochromatin in the cell nucleus.
Enhanced Transcriptional Capacity of the Non-Heterochromatic V296A Mutant
The selective failure of the V296A mutant to concentrate at pericentromeric heterochromatin, and its expression at the same level as the wild-type C/EBPα, results in a higher amount of C/EBPα in the non-pericentromeric subnuclear compartment where we previously showed active transcription to occur (31). Since the V296A mutant retained its ability to interact normally with some promoters (Fig. 3B), the enhanced amount of the V296A mutant in the active transcriptional compartment should result in a higher activation of those promoters by the V296A mutant than by equivalently expressed wild-type C/EBPα.
The C/EBP response element (C/EBP-RE) of the growth hormone promoter, to which wild-type and V296A mutant C/EBPα bound with equal affinity, was inserted in front of a minimal promoter (37). When transfected into pituitary progenitor GHFT1-5 cells, this reporter gene was activated on average, three to four-fold more by the V296A mutant than by the wild-type C/EBPα (Fig. 4A). A similar superactivation was observed at the PU.1 promoter (discussed later). There was minimal promoter activation by C/EBPα containing the V314P/V315P, L324P and L331P mutations in the leucine zipper (Fig. 4A) which, like the V296A mutant, are more concentrated in the non-pericentromeric compartment (Fig. 2B). In contrast to the V296A mutant, the leucine zipper mutants are more generally defective in binding at all DNA sites (Fig. 4B). The inactivity of these leucine zipper mutants showed that the enhanced activation by the V296A mutant did not originate indirectly from a simple excess of C/EBPα in the transcriptionally active compartment.
The V296A mutant superactivated transcription of the C/EBP-RE reporter in both GHFT1-5 cells (p=0.03) and undifferentiated 3T3-L1 cells (p=0.01) (Fig. 5A). Since this enhanced activation may have resulted from a heightened intrinsic transcriptional ability of the mutant C/EBPα, we compared the intrinsic the transcriptional activity the V296A mutant to wild-type C/EBPα. We replaced the leucine zipper (amino acids 312–358) of C/EBPα with a Gal4 DNA binding domain (DBD) and examined the ability of this C/EBP-Gal fusion protein to activate the expression of a reporter under the control of five adjacent Gal4 binding sites (Fig. 5B). The leucine zipper was deleted in these fusion proteins to disrupt dimerization and α-satellite DNA binding so that the WT and V296A Gal4 fusions are not targeted to the pericentromeric heterochromatin. In both 3T3-L1 and GHFT1-5 cells, the addition of wild-type or V296A C/EBPα sequences to the Gal4 DBD equivalently activated the Gal-responsive promoter. This provided strong evidence that the enhanced activation of the C/EBP-responsive promoter by the V296A mutant (Fig. 5A) was not due to an intrinsically higher transcriptional activation domain. Overall, this reporter activation data was consistent with the higher concentration of V296A in the non-heterochromatic subcompartment leading to enhanced promoter activation. As shown below, this conclusion is supported by promoter binding and activation studies in which C/EBPα binding to α-satellite DNA is reduced by either the V296A mutation or by the expression of Pit-1.
Enhanced Promoter Binding and Gene Activation by the V296A Mutant
The selective failure of the V296A mutant to bind α-satellite DNA would be expected to make V296A more available than wild-type C/EBPα for binding to an endogenous gene in the non-pericentromeric compartment of the cell nucleus. To examine this possibility, we examined the ability of the wild-type and mutant C/EBPα to bind to and activate a gene promoter. The PU.1 gene promoter was investigated because it is expressed in GHFT1-5 cells and contains a C/EBPα binding site that binds the V296A mutant with normal affinity (Fig. 3B).
We first compared the percentage of α-satellite and PU.1 promoter DNA precipitated by anti-C/EBPα antibodies in GHFT1-5 cells expressing wild-type or V296A mutant C/EBPα. Control cells expressing the vector alone were used as a negative control. Representative chromatin immunoprecipitations (ChIP) of α-satellite (Fig. 6A) or PU.1 promoter (Fig. 6B) DNA are shown. A series of bands that are multiples of the 234 bp long tandem repeated α-satellite DNA are detected when amplifying the α-satellite DNA (Fig. 6A). The amounts of α-satellite and PU.1 promoter DNA immunoprecipitated in four independent experiments were quantified and averaged (Figs. 6C). For the α-satellite DNA studies, the quantification was conducted on the monomeric, 234 bp band; the presence of the higher order bands indicates that the DNA precipitated included the tandem repetitive α-satellite DNA that predominates at pericentromeric heterochromatin.
The chromatin immunoprecipitations showed that wild-type C/EBPα associated with the α-satellite DNA in the cell (Fig. 6C, α-sat. ChIP, black bar). Thus, α-satellite DNA was available for binding by C/EBPα even though that DNA is present predominantly in presumably compact heterochromatin. The V296A mutant strongly reduced (p=0.03) the precipitation of α-satellite DNA (Fig. 6C, α-sat. ChIP, gray bar) confirming that, under the physiologic expression levels used, a reduced ability of V296A to bind α-satellite DNA in vitro correlated with the deficient association of this mutant with the heterochromatic regions. Interestingly, loss of α-satellite DNA binding was accompanied by a 2.7-fold increase (p=0.02) in binding of the V296A mutant to the PU.1 gene (Fig. 6C, PU.1 ChIP) and a corresponding 2.7-fold higher (p=0.002) amount of PU.1 transcripts in cells expressing the V296A mutant than in cells expressing wild-type C/EBPα (Fig. 6D). Thus, the selective inability of V296A C/EBPα to bind the α-satellite DNA resulted in an improved availability of V296A C/EBPα for binding to, and activation of, the PU.1 gene promoter.
Pit-1 Releases C/EBPα from Heterochromatin to Allow Promoter Binding and Activation
The ability of ectopically expressed Pit-1 to remove C/EBPα from pericentromeric heterochromatin in GHFT1-5 cells (35) provided a complementary method for manipulating the location of C/EBPα expressed in these cells. Pit-1 is a transcription factor that can synergize with C/EBPα at pituitary-specific promoters to which both C/EBPα and Pit-1 bind (34, 35, 37, 45–47). Consistent with the results of our V296A mutant studies, chromatin immunoprecipitation showed that co-expression of Pit-1 reduced the association of wild-type C/EBPα with α-satellite DNA (Fig. 7A, quantified from multiple experiments in Fig. 7C).
The functional consequences of Pit-1 removal of C/EBPα from α-satellite DNA and heterochromatin were assessed at the PU.1 promoter, which does not contain Pit-1 binding sites. Co-expression of Pit-1 led to increased binding of C/EBPα to the endogenous PU.1 promoter (Figs. 7B, C, p=0.02) and to a transfected PU.1 promoter (not shown, p=0.04). The Pit-1-mediated release of C/EBPα from α-satellite DNA, and enhanced PU.1 promoter binding, was accompanied by a 3.1-fold increase (p=0.001) in the amount of luciferase expressed from a co-transfected PU.1 promoter (Fig. 7D). Pit-1 by itself did not activate the PU.1 promoter, which reflects the absence of Pit-1 response elements in that promoter. Thus, Pit-1 expression enhanced C/EBPα binding to and activation of the PU.1 promoter.
If the enhanced C/EBPα activity observed upon Pit-1 expression was due predominantly to the enhanced concentration of C/EBPα in the non-heterochromatic compartment, then Pit-1 expression would not affect the activity of the already non-heterochromatic V296A mutant. Indeed, the 3.2-fold enhancement of PU.1 promoter activation caused by the V296A mutant was not significantly different from the 3.1-fold activation of wild-type C/EBPα resulting from Pit-1 expression (Fig. 7D). This is consistent with Pit-1 doing little more than removing C/EBPα from the heterochromatin. Furthermore, Pit-1 co-expression did not result in any further enhancement of the activity of the V296A mutant, which provided an experimental control demonstrating that there was no ‘classical’ cooperative activation by Pit-1 and C/EBPα in the current studies. Thus, the data are strongly consistent with an inhibition of C/EBPα activity by sequestration at pericentromeric heterochromatin that is reversed upon dispersal away from the heterochromatic compartment.
DISCUSSION
The data presented here demonstrate that the prevention of α-satellite DNA binding, either by Pit-1 co-expression or by the failure of the V296A mutant to bind α-satellite DNA, leads to release of C/EBPα from the heterochromatic subcompartment of the cell nucleus, a three-fold higher binding of C/EBPα to a promoter and a three-fold higher activation of that promoter. This data provides evidence that the subnuclear distribution of C/EBPα influences its effective concentration in the euchromatic domains of the cell nucleus. The results also demonstrate that it is the binding to the mouse major α-satellite DNA that sequesters this transcription factor at the heterochromatin.
The three-fold inhibition of C/EBPα activity through sequestration at pericentromeric heterochromatin is rather modest when compared to the strong inhibition of some transcription factors by their conceptually similar sequestration in the cytoplasm (48, 49). Nevertheless, we demonstrate that a transcription factor can be directly regulated by sequestration within the nucleus itself. This may lead to a general inhibition of, a fine-tuning of, or little effect on C/EBPα activity at gene promoters pertinent to biologic function. We speculate that the degree to which C/EBPα sequestration at heterochromatin regulates any specific promoter will be affected by the relative affinities of C/EBPα for the binding sites in that promoter and in the α-satellite DNA, and by the effect of chromatin architecture on the access of C/EBPα to each binding site.
The extent by which intranuclear sequestration contributes to biologic activity remains under investigation. The V296A mutant used to demonstrate the sequestration of transcriptional activity can not be used to assess directly the biologic relevance of C/EBPα sequestration since the V296A mutation also prevents C/EBPα binding to some sites and permits inappropriate C/EBPα binding to cyclic AMP response elements ((44) and unpublished data). This precludes the study of the V296A mutant, for instance, in mouse knock-in models. By contrast, some support for a biologic role of C/EBPα sequestration was indicated by prior studies showing that the ability of Pit-1 to re-locate C/EBPα away from the pericentromeric heterochromatin is disrupted by at least three point mutations in Pit-1 that are found in patients with combined pituitary hormone deficiency (34, 35, 47). Still, the biologic significance of the intranuclear sequestration of C/EBPα’s transcriptional activity remains to be firmly established.
Perhaps the strongest direct evidence of a biologic role for intranuclear sequestration of a transcription factor through binding to heterochromatic repetitive DNAs comes from studies in Drosophila where the pharmacologic disruption of repetitive DNA binding by the GAGA factor leads to homeotic transformations (20, 22). Furthermore, the treatment of mouse 3T3-442A cells with growth hormone, which induces adipocyte differentiation of these cells, results in a rapid and transient phosphorylation of the transcription factor C/EBPβ (a relative of C/EBPα); this modification is accompanied by a rapid and transient increase in the ability of C/EBPβ to bind to α-satellite DNA in vitro and an enhanced intranuclear association with pericentromeric heterochromatin (17). A number of other transcription factors also concentrate pericentromeric or non-pericentromeric sites within the cell nucleus (reviewed in (1, 23, 50, 51) and some of these distributions can vary under different physiologic conditions (10, 14–18, 20, 52). Therefore, transcription factor concentration at discrete domains within the cell nucleus may be somewhat commonplace but remains of unknown importance to biologic activity. The studies presented here provide the first steps towards understanding the significance of such events by formally demonstrating that transcriptional activity is indeed sequestered by the binding of a transcription factor to DNA sites concentrated in heterochromatin.
ACKNOWLEDGEMENTS
We thank Drs. Richard Day (University of Virginia) and Richard Weiner (U.C.S.F.) for critical reading of the manuscript.
The abbreviations used are
- C/EBPα
CCAAT/enhancer binding protein beta
- C/EBPα
CCAAT/enhancer binding protein alpha
- α-satellite DNA
mouse major alpha-satellite DNA
- EMSA
electrophoretic mobility shift assay
- PCR
polymerase chain reaction
- ChIP
chromatin immunoprecipitation
- C/EBP-RE
C/EBP response element
Footnotes
This work was supported by Public Health Service grant DK-54345 from the National Institutes of Health.
REFERENCES
- 1.Isogai Y, Tjian R. Curr Opin Cell Biol. 2003;15:1–8. doi: 10.1016/s0955-0674(03)00052-8. [DOI] [PubMed] [Google Scholar]
- 2.Kosak ST, Groudine M. Genes Dev. 2004;18:1371–1384. doi: 10.1101/gad.1209304. [DOI] [PubMed] [Google Scholar]
- 3.Misteli T. Cell. 2004;119:153–156. doi: 10.1016/j.cell.2004.09.035. [DOI] [PubMed] [Google Scholar]
- 4.Henikoff S, Ahmad K. Ann Rev Cell Dev Biol. 2005;21:133–153. doi: 10.1146/annurev.cellbio.21.012704.133518. [DOI] [PubMed] [Google Scholar]
- 5.Henikoff S, Ahmad K, Malik HS. Science. 2001;293:1098–1102. doi: 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- 6.Lamb JC, Birchler JA. Genome Biol. 2003;4 doi: 10.1186/gb-2003-4-5-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schueler MG, Higgins AW, Rudd MK, Gustashaw K, Willard HF. Science. 2001;294:109–115. doi: 10.1126/science.1065042. [DOI] [PubMed] [Google Scholar]
- 8.Perrod S, Gasser SM. Cell Mol Life Sci. 2003;60:2303–2318. doi: 10.1007/s00018-003-3246-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown KE, Guest SS, Smale ST, Hahm K, Merkenschlager M, Fisher AG. Cell. 1997;91:845–854. doi: 10.1016/s0092-8674(00)80472-9. [DOI] [PubMed] [Google Scholar]
- 10.Platero JS, Csink AK, Quintanilla A, Henikoff S. J Cell Biol. 1998;140:1297–1306. doi: 10.1083/jcb.140.6.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang QQ, Lane MD. Genes Dev. 1999;13:2231–2241. doi: 10.1101/gad.13.17.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown K. Crit Rev Eukaryot Gene Expr. 1999;9:203–212. doi: 10.1615/critreveukargeneexpr.v9.i3-4.50. [DOI] [PubMed] [Google Scholar]
- 13.Francastel C, Schübeler D, Martin DI, Groudine M. Nat Rev Mol Cell Biol. 2000;1:137–143. doi: 10.1038/35040083. [DOI] [PubMed] [Google Scholar]
- 14.Francastel C, Magis W, Groudine M. Proc Natl Acad Sci USA. 2001;98:12120–12125. doi: 10.1073/pnas.211444898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Georgopoulos K. Nat Rev Immunol. 2002;2:162–174. doi: 10.1038/nri747. [DOI] [PubMed] [Google Scholar]
- 16.Jolly C, Konecny L, Grady DL, Kutskova YA, Cotto JJ, Morimoto RI, Vourc'h C. J Cell Biol. 2002;156:775–781. doi: 10.1083/jcb.200109018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Piwien Pilipuk G, Galigniana MD, Schwartz J. J Biol Chem. 2003;278:35668–35677. doi: 10.1074/jbc.M305182200. [DOI] [PubMed] [Google Scholar]
- 18.Shestakova EA, Mansuroglu Z, Mokrani H, Ghinea N, Bonnefoy E. Nucl Acids Res. 2004;32:4390–4399. doi: 10.1093/nar/gkh737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cobb BS, Morales-Alcelay S, Kleiger G, Brown KE, Fisher AG, Smale ST. Genes Dev. 2000;14:2146–2160. doi: 10.1101/gad.816400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janssen S, Cuvier O, Muller M, Laemmli UK. Mol Cell. 2000;6:1013–1024. doi: 10.1016/s1097-2765(00)00100-3. [DOI] [PubMed] [Google Scholar]
- 21.Liu W, Enwright JF, Hyun W, Day RN, Schaufele F. BMC Cell Biol. 2002;3(1–11):6. doi: 10.1186/1471-2121-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Janssen S, Durussel T, Laemmli UK. Mol Cell. 2000;6:999–1011. doi: 10.1016/s1097-2765(00)00099-x. [DOI] [PubMed] [Google Scholar]
- 23.Corry GN, Underhill DA. Biochem Cell Biol. 2005;83 doi: 10.1139/o05-062. [DOI] [PubMed] [Google Scholar]
- 24.Blattes R, Monod C, Susbielle G, Cuvier O, Wu JH, Hsieh TS, Laemmli UK, Kas E. EMBO J. 2006;25:2397–2408. doi: 10.1038/sj.emboj.7601125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Darlington GJ, Wang N, Hanson RW. Curr Opin Genet Dev. 1995;5:565–570. doi: 10.1016/0959-437x(95)80024-7. [DOI] [PubMed] [Google Scholar]
- 26.Flodby P, Barlow C, Kylefjord H, Ahrlund-Richter L, Xanthopoulos KG. J Biol Chem. 1996;271:24753–24760. doi: 10.1074/jbc.271.40.24753. [DOI] [PubMed] [Google Scholar]
- 27.Inoue Y, Inoue J, Lambert G, Yim SH, Gonzalez FJ. J Biol Chem. 2004;279:44740–44748. doi: 10.1074/jbc.M405177200. [DOI] [PubMed] [Google Scholar]
- 28.Yang J, Croniger CM, Lekstrom-Himes J, Zhang P, Fenyus M, Tenen DG, Darlington GJ, Hanson RW. J Biol Chem. 2005;280:38689–38699. doi: 10.1074/jbc.M503486200. [DOI] [PubMed] [Google Scholar]
- 29.Joseph A, Mitchell AR, Miller OJ. Exp Cell Res. 1989;183:494–500. doi: 10.1016/0014-4827(89)90408-4. [DOI] [PubMed] [Google Scholar]
- 30.Stephanova E, Russanova V, Chentsov Y, Pashev I. Exp Cell Res. 1988;179:545–553. doi: 10.1016/0014-4827(88)90292-3. [DOI] [PubMed] [Google Scholar]
- 31.Schaufele F, Enwright JFI, Wang X, Teoh C, Srihari R, Erickson RL, MacDougald OA, Day RN. Mol Endocrinol. 2001;15:1665–1676. doi: 10.1210/mend.15.10.0716. [DOI] [PubMed] [Google Scholar]
- 32.Schaufele F, Wang X, Liu X, Day RN. J Biol Chem. 2003;278:10578–10587. doi: 10.1074/jbc.M207466200. [DOI] [PubMed] [Google Scholar]
- 33.Day RN, Periasamy A, Schaufele F. Methods. 2001;25:4–18. doi: 10.1006/meth.2001.1211. [DOI] [PubMed] [Google Scholar]
- 34.Day RN, Voss TC, Enwright JF, III, Booker CF, Periasamy A, Schaufele F. Mol Endocrinol. 2003;17:333–345. doi: 10.1210/me.2002-0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Enwright JF, III, Kawecki-Crook MA, Voss TC, Schaufele F, Day RN. Mol Endocrinol. 2003;17:209–222. doi: 10.1210/me.2001-0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schaufele F. Mol Endocrinol. 1999;13:935–945. doi: 10.1210/mend.13.6.0298. [DOI] [PubMed] [Google Scholar]
- 37.Schaufele F. J Biol Chem. 1996;271:21484–21489. doi: 10.1074/jbc.271.35.21484. [DOI] [PubMed] [Google Scholar]
- 38.Kummalue T, Friedman AD. J Leukocyte Biol. 2003;74:464–470. doi: 10.1189/jlb.1202622. [DOI] [PubMed] [Google Scholar]
- 39.Webb P, Nguyen P, Shinsako J, Anderson C, Feng W, Nguyen MP, Chen D, Huang SM, Subramanian S, McKinerney E, Katzenellenbogen BS, Stallcup MR, Kushner PJ. Mol Endocrinol. 1998;12:1605–1618. doi: 10.1210/mend.12.10.0185. [DOI] [PubMed] [Google Scholar]
- 40.Pfaffl MW. Nucleic Acids Res. 2001;29:2002–2007. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Landschulz WH, Johnson PF, Adashi EY, Graves BJ, McKnight SL. Genes Dev. 1988;2:786–800. doi: 10.1101/gad.2.7.786. [DOI] [PubMed] [Google Scholar]
- 42.Vinson CR, Sigler PB, McKnight SL. Science. 1989;246:911–916. doi: 10.1126/science.2683088. [DOI] [PubMed] [Google Scholar]
- 43.Nerlov C, Ziff EB. EMBO J. 1995;14:4318–4328. doi: 10.1002/j.1460-2075.1995.tb00106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller M, Shuman JD, Sebastian T, Dauter Z, Johnson PF. J Biol Chem. 2003;278:15178–15184. doi: 10.1074/jbc.M300417200. [DOI] [PubMed] [Google Scholar]
- 45.Chuang FM, West BL, Baxter JD, Schaufele F. Mol Endocrinol. 1997;11:1332–1341. doi: 10.1210/mend.11.9.9978. [DOI] [PubMed] [Google Scholar]
- 46.Jacob KK, Stanley FM. Endocrinol. 1999;140:4542–4550. doi: 10.1210/endo.140.10.7076. [DOI] [PubMed] [Google Scholar]
- 47.Demarco IA, Voss TC, Booker CF, Day RN. Mol Cell Biol. 2006;26:8087–8098. doi: 10.1128/MCB.02410-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vandromme M, Gauthier-Rouviere C, Lamb N, Fernandez A. Trends Biochem Sci. 1996;21:59–64. [PubMed] [Google Scholar]
- 49.Komeili A, O'Shea EK. Curr Opin Cell Biol. 2000;12:355–360. doi: 10.1016/s0955-0674(00)00100-9. [DOI] [PubMed] [Google Scholar]
- 50.DeFranco DB. Mol Endocrinol. 2002;16:1449–1455. doi: 10.1210/mend.16.7.0880. [DOI] [PubMed] [Google Scholar]
- 51.Hager GL, Lim CS, Elbi C, Baumann CT. J Steroid Biochem Mol Biol. 2002;74:249–254. doi: 10.1016/s0960-0760(00)00100-x. [DOI] [PubMed] [Google Scholar]
- 52.Tang QQ, Lane MD. Proc Natl Acad Sci USA. 2000;97:12446–12450. doi: 10.1073/pnas.220425597. [DOI] [PMC free article] [PubMed] [Google Scholar]