Abstract
The suprachiasmatic nucleus (SCN) of the hypothalamus is the site of the master circadian pacemaker in mammals. The individual cells of the SCN are capable of functioning independently from one another and therefore must form a cohesive circadian network through intercellular coupling. The network properties of the SCN lead to coordination of circadian rhythms among its neurons and neuronal subpopulations. There is increasing evidence for multiple interconnected oscillators within the SCN, and in this Review, we will highlight recent advances in our understanding of the complex organization and function of the cellular and network-level SCN clock. Understanding the way in which synchrony is achieved between cells in the SCN will provide insight into the means by which this important nucleus orchestrates circadian rhythms throughout the organism.
THE CIRCADIAN NETWORK
Behavioral and physiological processes are coordinated by endogenous biological clocks, permitting organisms to synchronize to and anticipate changes in the environment. The circadian system serves this purpose by generating and sustaining 24h rhythms of biological processes and by synchronizing to external stimuli such as the solar day-night cycle. In mammals, circadian clocks exist throughout the body, in individual cells and organs, and these clocks are kept synchronized by a master pacemaker residing in the suprachiasmatic nucleus (SCN) of the hypothalamus [1, 2] (Glossary). The SCN, however, is potentially comprised of ~20,000 cell autonomous clocks of its own [3, 4]. Single neurons within the SCN exhibit independent rhythms of firing rate [4] and gene expression [5]. Advances in electrophysiological and imaging techniques, allowing long-term, single-cell recording of circadian rhythmicity, have improved our understanding of the role of both the cell autonomous clock and the SCN network in maintaining a stable circadian system. In addition, recent work has demonstrated that the connectivity between these individual SCN neurons is not only essential for proper timekeeping, but also imparts properties that distinguish the SCN as a master pacemaker of the circadian clockwork. Here, we focus on the network formed by the independent cellular oscillators of the SCN. We will review the evidence for multiple oscillators within the SCN, highlighting recent advances in our understanding of the complex organization and function of the cellular and network-level SCN clock. We will examine the importance of the SCN oscillator network for the robustness of the circadian system and discuss current views on the mechanism and function of coupling between SCN neurons.
THE SUPRACHIASMATIC NUCLEUS
The SCN is a heterogenous structure (Figure 1; see [6] for a comprehensive review). Subregions can be defined throughout the nucleus based on peptidergic content, anatomical location, and circadian parameters of the component cells. Traditionally, the SCN has been divided into two major subdivisions known as the dorsomedial (dmSCN) “shell” and the ventrolateral (vlSCN) “core” [3, 7]. The former contains cells expressing arginine vasopressin (AVP), while the latter is characterized by expression of vasoactive intestinal polypeptide (VIP), gastrin-releasing peptide (GRP) [3, 8], and neuromedin S (NMS) [9, 10] (Figure 1). There are numerous projections from the core to the shell, but fewer efferents from the dorsal shell innervate the ventral core [11]. Although they have served as a useful framework for exploring intercellular communication within the master pacemaker, the anatomically- and peptidergically-derived classifications of dm/vlSCN (or core/shell) do not begin to capture the complexity of the nucleus (see [12, 13] for further discussion). Variability exists along every axis of the SCN [14], demanding a rigorous examination of the function of individual SCN oscillators and subregions in the control of mammalian circadian rhythms.
FIG 1. Expression of neuropeptides in the mammalian SCN.

Photomicrographs of mouse SCN slices. The bilateral nuclei are located directly above the optic chiasm and positioned on either side of the 3rd ventricle. Slices were obtained from transgenic mice expressing the CLOCKΔ19 mutant protein tagged with hemagglutinin (HA) in secretogranin positive cells (expression of the HA-tagged transgene protein product is shown in green). Sections were immunostained to detect expression of the neuropeptides (A) AVP (red) and (B) VIP (red). AVP expression is observed most prominently in the dorsal SCN, whereas VIP expression is strongest in the ventral SCN. Cells expressing both CLOCKΔ19-HA (expressed throughout the nucleus) and the peptide of interest appear in yellow. Reproduced, with permission, from [113].
Within most (if not all) mammalian cells, a transcriptional-translational feedback loop keeps circadian time (see [15, 16] for review; Figure 2). The basic-helix-loop-helix transcription factors CLOCK and BMAL1 accumulate and heterodimerize in the nucleus. Acting as a positive regulator, the CLOCK:BMAL1 complex activates transcription of Period (Per) and Cryptochrome (Cry) genes, leading to the subsequent accumulation of PER and CRY proteins. As their levels increase in the cytoplasm, PER and CRY dimerize, re-enter the nucleus, and the complex suppresses transcriptional activation by CLOCK and BMAL1. In addition to this core feedback loop, there are accessory loops in the molecular clock. The CLOCK:BMAL1 complex activates Rev-erbα. Rev-erbα is a transcriptional repressor of Bmal1, providing a mechanism by which buildup of BMAL1 protein can suppress further Bmal1 transcription. Eventually, PER and CRY proteins are degraded, at least in part through E3-ligase-mediated proteosomal degradation [17-21]. Casein kinase 1 (CK1) isoforms (α, δ and ε) contribute to protein turnover by phosphorylating PER, targeting it for degradation [22-26]. How the cell autonomous molecular clock drives biological and physiological circadian output rhythms is only poorly understood. However, it is clear that the coupling among oscillators within the SCN is vital to maintaining robust circadian rhythmicity.
FIG 2. Schematic of the intracellular molecular clock and model of intercellular signaling pathways.
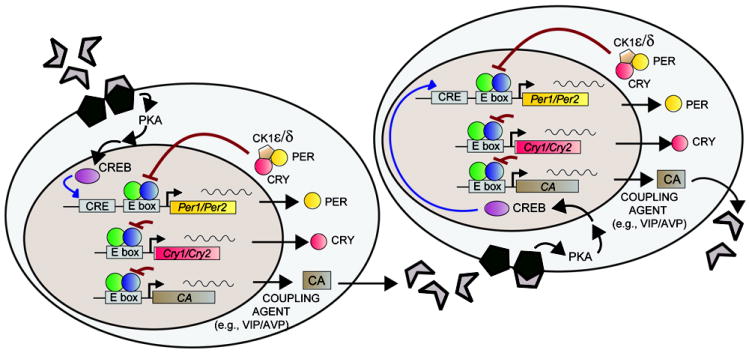
The molecular circadian clock consists of the positive elements CLOCK and BMAL1 (represented by the green and blue circles, respectively), which dimerize and activate transcription of Per and Cry genes. PER and CRY dimerize and translocate to the nucleus to suppress further transcriptional activation by CLOCK:BMAL1. CK1 regulates protein turnover, contributing to the degradation and nuclear translocation of PER. Intercellular coupling agents (CA), which may include VIP, AVP, or other neuropeptides, activate cAMP-signaling pathways [including protein kinase A (PKA)]. These signaling cascades result in CREB-dependent transcriptional activation of Per genes. Adapted, with permission, from [79].
THE PHOTIC ENVIRONMENT AND SCN OSCILLATORS
Constant Light Reveals Multiple Circadian Oscillators within the SCN
Perhaps the first evidence that the mammalian circadian system was comprised of multiple oscillators at the organismal level was the observance of “splitting” of locomotor activity rhythms in the ground squirrel (Spermophilus undulatus) [27] and golden hamster (Mesocricetus auratus) [28]. Hamsters housed in constant light display two independent rhythmic components of locomotor activity [28]. The two activity components are associated with independently oscillating rhythms of clock gene expression in the right and left SCN [29, 30], demonstrating that these bilateral nuclei are capable of functioning independently of one another. Housing hamsters and other nocturnal rodents in constant bright light for extended durations of time results in a lengthening of the circadian free-running period and, in some cases, behavioral arrhythmicity [31-33]. This phenomenon is likely due to decoupling of neurons within the SCN [34]. Decreased amplitude of rhythmicity of neuronal firing rate [30] and clock gene expression in the SCN as a whole [33, 34], and a wider range of peak phases within individual SCN neurons [34], can be observed in animals that have been housed in constant light. The rhythmicity of individual cells, however, remains robust [34]. The behavioral disruptions occurring in constant light are a product of oscillator desynchrony within the SCN, as opposed to a defect in cell-autonomous circadian rhythmicity [34].
SCN Oscillators Differ in Their Response to Changes in Photic Environment
Jetlag paradigms, in which the environmental light:dark cycle has been advanced or delayed by several hours, have revealed distinct resetting properties of anatomical subregions within the SCN. Both in situ hybridization and luciferase reporter studies have demonstrated that, in response to a large shift in the light:dark cycle, molecular circadian rhythms shift rapidly in the vlSCN, with rhythms in the dmSCN taking longer to respond to the change in photic environment [35, 36]. This results in transient internal phase desynchronization within the SCN. It has been hypothesized that the retino-recipient vlSCN is immediately responding to the photic phase advance and communicating this information to the dmSCN [36]. The dmSCN may then modulate the response of the vlSCN, with coupling between subregions eventually achieving an appropriate phase throughout the nucleus [36]. The balance between intra-SCN autonomy and connectivity could explain the gradual shift in behavior and physiology, and temporary internal desynchronization [37], that occurs following rapid changes in the light:dark cycle. In addition to differences in the kinetics of phase-shifting between the dm and vlSCN, differences in the peak expression of clock genes and the rate of resetting can be observed along the rostral-caudal extent of the SCN [36], further emphasizing the complexity of the SCN oscillator network.
Regional changes in SCN clock gene expression and neuronal firing are also seen in response to changes in photoperiod (simulating seasonal changes in day length). Long days result in a broadened pattern of SCN clock gene expression [38-41] and neuronal activity [42, 43]. Compared to short days, more widely distributed phases of peak neuronal activity among subpopulations of SCN neurons are observed during long days [42, 43]. Day length induced changes in the phase and waveform of rhythms vary across both the dorsal-ventral and rostral-caudal extents of the SCN [38, 40, 44].
Changing the length of the photic cycle can also desynchronize SCN oscillators [45-49]. By housing rats (Rattus norvegicus) under 22h light:dark schedules (11h light/11h dark), as opposed to the typical 24h day, molecular rhythms in the dm and vlSCN can dissociate [46]. Distinct rhythms of clock gene expression can be observed, with the vlSCN remaining entrained (ie. synchronized) to the light:dark cycle [46]. The dmSCN, however, is only relatively coordinated to the light:dark cycle under these conditions, displaying bouts of free-run [46, 49]. Behaviorally, rats exhibit two components of locomotor activity with different period lengths, presumably emerging from the outputs of now uncoupled SCN subregions [46]. Body temperature and slow wave sleep also exhibit two distinct circadian components under this forced desynchrony protocol [45], whereas the rhythm of rapid eye movement (REM) sleep preferentially follows the dmSCN [47]. These findings not only confirm that rhythms in the dm and vlSCN are capable of functioning independently, but suggest that these two oscillatory subregions can independently influence rhythms throughout the organism. The way in which light-entrained oscillators located within the vlSCN are able to couple to oscillators throughout the SCN to maintain stable entrainment has yet to be characterized. Presumably, under conditions of normal entrainment to a light:dark cycle, coupling among the many oscillators of the SCN network must occur to orchestrate robust rhythmicity among all rhythmic biological processes.
COUPLING WITHIN THE SCN
In contrast to what is observed following jetlag or under conditions of forced desynchrony, the multiple oscillators of the SCN normally function in a unified manner. However, even in the coordinated, intact SCN, cellular oscillations are not completely synchronous. Individual neurons exhibit a range of phases [50]. Heterogeneity of phase can be observed along every anatomical dimension of the SCN [14]. There is a “wave” of oscillatory activity within the SCN [14, 36, 51]. Expression of Per genes and their protein products appears to spread through the nucleus, with the earliest peaking populations of neurons observed in the medial and dorsal SCN [14, 36, 51-53]. This pattern of PER expression within SCN subregions occurs in a consistent, stable manner, displaying both temporal and anatomical organization [14, 53] (Figure 3). The phase of subpopulations of SCN neurons does not necessarily correlate with the circadian period or peptidergic content of the cells [14]. The process by which this oscillatory pattern is organized within the SCN network is unknown. Determining the mechanism that establishes the phase of individual neurons and populations of neurons, and the hierarchy of SCN neuronal connectivity, will be a critical step towards understanding the SCN network. In-depth analysis of the spatiotemporal organization of the SCN [14, 53, 54] is likely to provide insight into regulation of the SCN itself and circadian outputs.
FIG 3. Organized PER2 expression in the SCN.

Assessment of luciferase bioluminescence in acute brain slices of the SCN from PER2∷LUC mice using unconstrained cluster analysis [53]. The analysis reveals a wave of oscillatory activity within the SCN, with expression of PER2 (as assessed by the bioluminescence signal of the LUC reporter gene) spreading through the nucleus. This pattern of serial activation of PER2 can be visualized as a “map” of distinct SCN subregions, defined here solely by the spatiotemporal distribution of the bioluminescent signal, without any underlying assumptions being specified by the experimenter. (A) The color coded ‘map’ indicates the location of each cluster in the slice. Note the substantial symmetry between the left and right sides of the SCN. (B) The time series graph displays the mean brightness for each distinct cluster over time [53]. Such a finding reveals that circadian oscillation is characterized by a stable daily cycle of gene expression involving orderly serial activation of specific SCN subregions, followed by a silent interval [53]. Time series for clusters in the map that cannot be discerned from scatter of light are not shown. Figure provided by the authors. Adapted, with permission, from [53].
When SCN cells are dispersed in culture, the observed phases and periods are much more variable and the rhythms of individual neurons are significantly less reliable [4, 55-57], although synchrony can be observed when dispersed neurons are cultured at a high density [58]. These findings support the notion that the anatomical organization of the SCN and cell-to-cell communication permits coupling between autonomous cell oscillators to convey coordinated rhythms within the nucleus.
NETWORK PROPERTIES CONFER STABILITY AND ROBUSTNESS TO THE SCN AND THE CIRCADIAN SYSTEM
In addition to maintaining synchrony within SCN neuronal populations, there are two other important functions of SCN coupling which have come to light over the past several years: 1) providing robustness, precision, and stability to SCN rhythms; and, 2) generating and sustaining circadian rhythms.
Coupling of SCN neurons leads to robustness (strength and precision of rhythmicity, as well as resistance to perturbation) of the SCN itself and its output rhythms. As discussed above, the range of periods of individual, widely-dispersed cultured SCN neurons is greater and less stable than that of neurons recorded from whole SCN explants. This suggests that, in the intact nucleus, coupling between neurons serves not only to provide local synchrony, but also to influence the period of oscillations within individual cells and determine the period of the SCN as a whole. Indeed, it has been demonstrated that the period of locomotor behavior of hamsters can be estimated by averaging the period of neuronal firing rate of single cells within the SCN [59]. Mathematical modeling demonstrates that synchronization to the mean circadian period occurs once intercellular coupling strength reaches a certain threshold [59]. Thus, in widely dispersed culture systems, coupling strength is reduced resulting in only partial synchronization and a broad range of periods among individual cells [59]. The contribution of coupled oscillators to robustness of period is also observed in the mouse. There is less variability in the period of mouse (Mus musculus) locomotor activity and SCN explant Per1 expression than in cellular-level firing rate [55]. Even the most stable dispersed individual cells are only as precise as the least stable mouse locomotor rhythm or SCN explant [55]. Coupling of variable cellular oscillators results in the high amplitude, stable rhythms observed at the tissue, system, and organismal levels.
The SCN oscillator network also conveys robustness against genetic mutations. Animals with chimeric SCN, comprised of both wildtype and circadian mutant (either the short period Tau mutation or the long period ClockΔ19 mutation) tissue, exhibit behavioral rhythms with components intermediate to, and sometimes dually expressing, the periods of the component cells [60, 61]. Neurons from the SCN of ClockΔ19 mice (which posses a dominant negative form of CLOCK, resulting in a lengthened circadian period and eventual arrhythmicity when housed in constant lighting conditions) maintain robust rhythmicity of firing rates in the intact nucleus [62]. However, in cell dispersals only a small percentage of SCN neurons from ClockΔ19 mice are able to maintain circadian firing rate rhythms [62]. Deletion of Per1 or Cry1 results in impaired rhythmicity in dissociated SCN neurons and fibroblasts, with many cells exhibiting no measurable rhythm [5]. The circadian rhythms of SCN explants and of behavior do not reflect this cell-autonomous loss of rhythmicity. SCN explants from mice null for either Per1 or Cry1 remain rhythmic, as does the locomotor activity of the animals [5]. This result suggests that coupling between (largely arrhythmic) individual neurons in the SCN slice (and in the mouse) are able to compensate for loss of cell autonomous rhythmicity [5] (Figure 4).
FIG 4. Intercellular coupling compensates for genetic deficits.
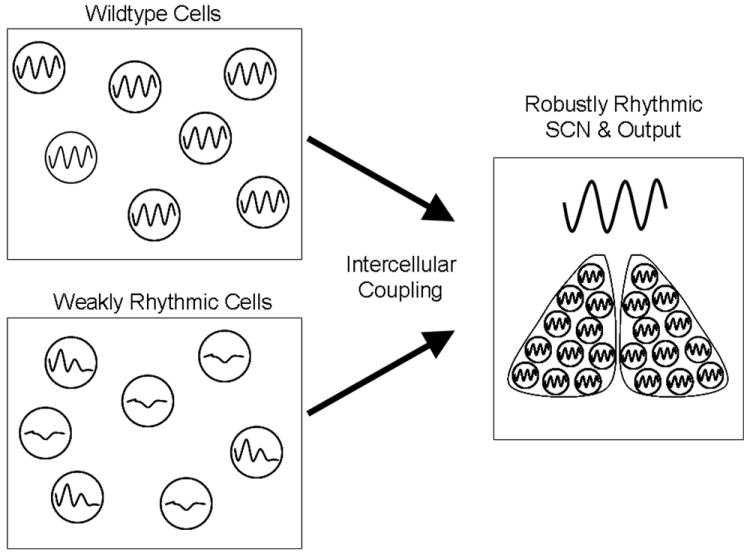
Uncoupled SCN cells are susceptible to arrhythmicity due to genetic defects in the molecular clock. Dispersed SCN cells from mice null for either Per1 or Cry1 display weak rhythms [5] SCN explants from these animals, however, are rhythmic, as is their locomotor activity [5]. Coupling of weakly rhythmic cellular oscillators can compensate for compromised circadian function, preserving stable rhythmicity at the level of the SCN and output rhythms. Individual cells derived from wildtype animals are rhythmic, although increased arrhythmicity and variability in period is observed in widely dispersed cultures [4, 55]. Thus, even among independent cells that are already rhythmic, coupling serves to stabilize the rhythmicity of the SCN.
Recently, it was demonstrated that network properties of the SCN impart resistance to temperature resetting. Temperature is a powerful entraining cue for many species [63-65], but in mammals and other homeotherms the entraining effects of temperature are less robust, with temperature cycles serving as weak entraining signals (at best) in rodents [66-68] and primates [69]. Temperature entrainment of circadian gene expression rhythms has been observed in mammalian peripheral cells and tissues, but the SCN itself is less sensitive to temperature cues [70-72]. While mouse pituitary and lung explants entrain to daily temperature cycles, the SCN free-runs through a 24 h (12 h 36°C: 12 h 38.5°C) temperature cycle, seemingly uninfluenced by temperature cues [72]. When communication between SCN neurons is blocked with tetrodotoxin (TTX, which blocks voltage-gated sodium channels) or nimodipine (an L-type calcium channel blocker), however, temperature pulses synchronize and reset rhythms in individual cells resulting in a temperature-sensitive SCN [72] (Figure 5). Similarly, lung, but not SCN explants, entrain to temperature cycles of 20h (10h 35.5°C: 10h 37°C) and 28h (14h 35.5°C: 14h 37°C) [70]. Only large changes in temperature (≥ 6°C), cycling with a more moderate period (22h), are able to entrain PERIOD2 (PER2) rhythms in the SCN [70] (as measured in PER2∷LUC mice, which express PER2 fused with the luciferase [LUC] reporter [73]). These experimental findings have been supported by theoretical models demonstrating that coupling within an oscillator network (eg., the SCN) limits the range of entrainment [70].
FIG 5. Network properties convey temperature resistance to the SCN.

(a) Bioluminescence trace from an SCN explant obtained from PER2∷LUC mice (bioluminescence represents PER2∷LUC expression). The SCN is resistant to perturbation by temperature signals. Blocking communication among SCN neurons with TTX allows a 6h temperature pulse (temperature increased to 38.5°C, indicated by yellow bar) to reset the phase of the SCN. (b) Phase transition curves (PTCs) plotting phase prior to temperature pulse (“old phase”) against phase following temperature pulse (“new phase”). PTCs are from SCN cultures from PER2∷LUC mice without drug (gray), with 1μM TTX (red), and with 5μM TTX (blue). Following TTX treatment, the SCNs display type 0 phase-resetting. Type 0 resetting is strong resetting, with oscillators resetting to a common phase, regardless of the phase at which the resetting signal is administered. (c) Heat maps demonstrating the effects of TTX on temperature resetting in individual neurons within an SCN culture. In whole SCN explants, phase and period length are stable and robust, although some variability in phase can be observed (left). Following TTX treatment, SCN neurons are uncoupled (right). Individual neurons in the TTX-treated condition free-run, resulting in a wide range of phases. TTX also renders individual SCN neurons sensitive to temperature-induced phase resetting. A 6h temperature pulse (temperature increased to 38.5°C, indicated by yellow bar) synchronizes the phases of individual neurons in the TTX-treated SCN. No effect on phase was observed when a temperature pulse was administered in the absence of TTX. Heat maps were generated by imaging single cell-sized regions of interest using a cooled charge-coupled device (CCD) camera. Maps display voxels measured from dorsal to ventral through the SCN. Red corresponds to the peak of bioluminescence and green to the trough. Reproduced, with permission, from AAAS [72].
Together, these data demonstrate that coupling within the mouse SCN conveys resistance to temperature entrainment and imparts robustness to circadian rhythmicity within the nucleus. Such resistance to temperature resetting may allow the SCN to control peripheral oscillators through body temperature rhythms while itself remaining unaffected by this rhythmic output [72]. This hypothesis needs to be directly tested on the background of a coupled and uncoupled SCN network. The data discussed above conflict with previous findings that the rat SCN can be phase-shifted [74] and entrained [75] by temperature. It is possible that the discrepancy between studies is due to a species difference in the sensitivity of the SCN to temperature signals. Such species differences in SCN resetting could result from underlying differences in SCN coupling strength or circadian amplitude (as seen with the ClockΔ19 mutation [76]). A more careful comparison of the effect of temperature on SCN explants from a variety of mammalian species is warranted. Interestingly, in one report of temperature sensitivity in the rat SCN, explants were taken from neonatal animals [75]. This raises the possibility that developmental changes in intercellular signaling may occur which confer temperature-resistance and other network attributes to the SCN. Investigating the developmental contributions to SCN function may reveal new properties of cellular communication within the oscillator network.
INTERCELLULAR SIGNALING PLAYS A ROLE IN CIRCADIAN RHYTHM GENERATION
Beyond sustaining circadian rhythmicity, there is recent evidence that network properties of the SCN are capable of generating rhythmicity. Mathematical models have demonstrated that intercellular coupling can sustain rhythmicity in individual cellular oscillators, and experimental evidence supports this prediction [5, 77]. Culturing SCN neurons at very low densities results in a substantial decrease (as compared to explants or high density cell dispersals) in the number of neurons exhibiting sustained circadian rhythms of firing rate and PER2 expression [78], suggesting that coupling between SCN neurons is necessary to sustain rhythmicity in individual cells. The experiments in cells from Per1-/- and Cry1-/- mice demonstrate that even a few intrinsically rhythmic neurons are capable of producing circadian oscillations throughout the SCN, an effect which is also achieved through intercellular coupling [5, 62]. Surprisingly, rhythmic protein expression can also be observed from an SCN containing no intrinsically rhythmic neurons. Highly variable, rhythmic oscillations in PER2∷LUC have recently been observed from the SCN of Bmal1-/- mice, which are behaviorally arrhythmic [79]. SCN explants from Bmal1-/- mice display PER2∷LUC rhythms with periods ranging from 14-30h. While there is stochastic variability in the period length of oscillations from Bmal1-/- SCN explants, these unstable rhythms persist for over 30 cycles in culture. Dissociated cells from the SCN of these mice possess no rhythmic PER2 expression, although PER2 levels fluctuate in a random manner. The rhythms observed in SCN explants, therefore, appear to be an emergent network property resulting from intercellular coupling on a background of molecular noise. Indeed, SCN-level oscillations are abolished by uncoupling SCN neurons with TTX [79]. Mathematical modeling provides theoretical confirmation that coupling among arrhythmic SCN neurons, which demonstrate fluctuating but not oscillating PER2 levels, can produce tissue-level rhythmicity [79]. This work demonstrates that the SCN network is capable of generating oscillations in the circadian range from a population of individually arrhythmic cells, illustrating a previously unappreciated level of robustness in the SCN system.
MECHANISMS OF INTERCELLULAR COMMUNICATION WITHIN THE SCN
The mechanisms by which individual cells in the SCN couple to produce stable, coherent rhythms are not fully understood. As discussed above, blocking sodium-dependent action potentials with TTX [50, 79] or isolating neurons within the SCN [78] leads to cellular desynchrony. This suggests that neurotransmission is vital to maintenance of circadian rhythmicity, although gap junctions may also play an important role in SCN coupling [80, 81].
Evidence for the involvement of neurotransmitters in SCN neuronal coupling
Considerable effort has been made to identify the signals responsible for coupling within the SCN network, with several of the prominent SCN neurotransmitters, includingVIP, GRP, and GABA, being prime candidates. The strongest support for a neuropeptide coupling agent within the SCN has been put forth for VIP (see [82, 83] for reviews). VIP modulates light-induced phase-resetting [84], and VIP application shifts the phase of both the SCN (in vitro) [85, 86] and locomotor activity [87]. Mice deficient in VIP or its receptor, VPAC2 (encoded by the gene Vipr2), have weak (or disrupted) locomotor activity rhythms [58, 88, 89]. Loss of VIP-signaling results in desynchrony among SCN neurons [90-92]. Individual SCN neurons from VIP and VPAC2 null mice are frequently arrhythmic, both in dispersed culture and in organotypic explants, and lower amplitude rhythms are observed among the remaining rhythmic cells [58, 90-92]. Interestingly, variability in phase among SCN neurons from both Vip-/- and wildtype mice is negatively correlated with the robustness of behavioral rhythms [93]. In the in vivo condition, it is likely that VIP (among other neuropeptidergic signals) provides an intercellular coupling signal for SCN oscillators, stabilizing and synchronizing rhythms among SCN neurons.
While there is ample evidence for the crucial role of VIP-signaling in SCN neuronal coupling, other neurotransmitters are likely involved. GRP and GABAA receptor activation both produce phase-shifts in SCN oscillations and locomotor activity rhythms [87, 94-97]. Application of GRP or GABA can synchronize asynchronous SCN neurons [92, 95]. However, there is also evidence that GABA antagonism can actually improve stability and precision in circadian firing rate of SCN neurons [98]. Further research is needed to determine the way in which these neurotransmitter systems interact at the cellular level to impart tissue-level synchrony and stability to the SCN.
Calcium Signaling May Underlie Intracellular Responses to Oscillator Coupling
Calcium signaling plays an important role in the SCN network. cAMP activation induces Per transcription [99] and inhibiting cAMP signaling in organotypic SCN slices results in decreased circadian amplitude [100]. Activation of cAMP/cAMP response element-binding protein (CREB)-dependent pathways are involved in light-induced resetting of the SCN [101, 102], with CREB repression resulting in an aberrant response to photic stimuli and reduced PER expression [103]. This pathway is also an important target of VIP signaling. VIP receptor activation results in increased intracellular cAMP [86, 104, 105] and, within the SCN, VIP has phase-dependent effects on intracellular calcium levels [106]. These effects of VIP are likely due to activation of adenylate cyclase and phopholipase C pathways [86, 107]. Thus, transcriptional activation of Per1 through CREB-dependent pathways is likely to confer the synchronization properties of VIP, and perhaps other neuropeptides, within the SCN. Interestingly, the stochastic rhythmicity observed in SCN explants from Bmal1-/- mice is lost upon application of cAMP inhibitors [79], providing further evidence that cAMP/CREB-mediated transcriptional regulation is necessary for intercellular coupling. The existing data are consistent with the hypothesis that a number of neurotransmitter and signaling pathways may converge upon CREB to integrate intercellular signaling into molecular changes at the level of an individual cellular clock (Figure 2).
The calcium-activated potassium (BK) channel also plays a role in SCN function. BK channel null mice display a decreased circadian amplitude of body temperature [108] and locomotor activity [108, 109]. Interestingly, the effect of BK channel deletion on circadian output rhythms does not appear to be a consequence of alterations in the molecular clock; rhythms of clock gene expression remain unchanged [108]. The neuronal firing rate of the SCN, however, is affected, with dampened rhythmicity observed in BK channel null mice [108, 109]. The effect of BK channel loss on neuronal firing rate occurs at the level of individual neurons [108], and a specific role for BK channels in intercellular synchrony has not yet been identified [109] (although this cannot be ruled out). It is not known how this change in cell-level firing rate is integrated at the network-level to contribute to the output of the SCN or the behavioral phenotype of the mice.
UNRAVELING THE SCN NETWORK
Intercellular coupling of cell autonomous oscillators within the SCN renders synchrony and robustness to the master circadian pacemaker. Network level properties convey a stable circadian phase and period at the tissue level to a nucleus comprised of intrinsically variable neuronal rhythms. The ability of this complex network to generate rhythmicity upon a background of arrhythmicity [79] provides yet another piece of evidence for the importance and intricacy of coupling within the SCN.
While our knowledge of the role of networking in SCN function has advanced significantly in recent years, our grasp of the fundamental components of the network remains lacking (Box 1). Combining mathematical simulations of the oscillator network with experimental approaches [5, 70, 79, 110] will likely be necessary to unravel the complexity of SCN intercellular connectivity and the output of this multi-oscillator pacemaker. Moreover, the effect of silencing of individual neurochemical and anatomical components of the SCN network should be examined. In the fly (Drosophila melanogaster), studies using transgenic models and targeted ablation of specific populations of clock neurons have revealed mechanistic details of circadian network connectivity [111, 112]. Similar investigations should be undertaken in mammals, using emerging tools such as peptide-specific neurotoxins and cell-type specific cre-recombinase expressing mice. Deciphering the intercellular circadian code is an important step in advancing the study of the SCN network. Understanding the way in which balance is maintained between cell autonomy and synchrony within the SCN, and the manner in which this is translated into output signals, will provide insight into the means by which this nucleus coordinates a complicated web of circadian processes throughout the organism.
Box 1. Outstanding Questions.
What are the intercellular communication signals within the SCN?
How do intercellular coupling signals get transmitted into intracellular changes in neuronal oscillator function?
Is there a role for non-transcriptional/translational components of the cellular clock in intercellular coupling of circadian oscillations? Do cytosolic rhythms constitute a critical feature of the network properties of the SCN?
How are signals from individual cellular oscillators integrated?
What mechanisms allow the SCN network to simultaneously drive circadian outputs with varied phases?
How are SCN output signals formed?
What are the specific roles of calcium-signaling and calcium-sensitive BK potassium channels in the SCN network?
How do signals from the SCN control behavior and other processes? What are the relative contributions of humoral factors and sympathetic innervation?
Do similarly intricate oscillator networks exist outside of the SCN? Are these networks important for sustaining rhythms in their own component cells?
Acknowledgments
We thank Drs. Rae Silver and Nicholas Foley for the contribution of Figure 3. We also thank Dr. Seung-Hee Yoo for helpful discussions. This work was supported by the National Institutes of Health (NIH P50 MH074924 and R01 MH078024 to J.S.T). J.S.T. is an Investigator in the Howard Hughes Medical Institute.
Glossary
- AVP
Arginine vasopressin, a neuropeptide that is widely expressed in the dmSCN [3, 8] and exhibits a circadian pattern of release [114, 115]
- Entrainment
synchronization of an oscillation to an environmental stimulus occurring at regular intervals (usually ~24h)
- Free-running period
the period of an oscillation in the absence of any entraining signals. The free-running period reflects the intrinsic period of the oscillator, uninfluenced by environmental timing cues
- GRP
Gastrin-releasing peptide, highly expressed within neurons of the vlSCN [3]. GRP administration results in phase-shifts of behavioral and SCN rhythms [87, 94, 96]
- SCN
The suprachiasmatic nucleus is the site of the master circadian pacemaker in mammals. The SCN consists of two, bilateral nuclei positioned above the optic chiasm and on either side of the 3rd ventricle. The mouse SCN has been estimated to contain ~20,000 neurons [3]
- VIP
Vasoactive intestinal polypeptide, highly expressed within the vlSCN [3, 8]. The VIP receptor VPAC2 (encoded by Vipr2) is expressed both within the VIP-ergic neurons as well as in the cells of the dmSCN [116]. VIP signaling is important in the maintenance of intercellular synchrony within the SCN
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Moore RY, Eichler VB. Loss of a circadian adrenal corticosterone rhythm following suprachiasmatic lesions in the rat. Brain Res. 1972;42:201–206. doi: 10.1016/0006-8993(72)90054-6. [DOI] [PubMed] [Google Scholar]
- 2.Stephan FK, Zucker I. Circadian rhythms in drinking behavior and locomotor activity of rats are eliminated by hypothalamic lesions. Proc Natl Acad Sci U S A. 1972;69:1583–1586. doi: 10.1073/pnas.69.6.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abrahamson EE, Moore RY. Suprachiasmatic nucleus in the mouse: retinal innervation, intrinsic organization and efferent projections. Brain Res. 2001;916:172–191. doi: 10.1016/s0006-8993(01)02890-6. [DOI] [PubMed] [Google Scholar]
- 4.Welsh DK, et al. Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms. Neuron. 1995;14:697–706. doi: 10.1016/0896-6273(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 5.Liu AC, et al. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell. 2007;129:605–616. doi: 10.1016/j.cell.2007.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Welsh DK, et al. Suprachiasmatic nucleus: cell autonomy and network properties. Annu Rev Physiol. 2010;72:551–577. doi: 10.1146/annurev-physiol-021909-135919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moore RY. Organization and function of a central nervous system circadian oscillator: the suprachiasmatic hypothalamic nucleus. Fed Proc. 1983;42:2783–2789. [PubMed] [Google Scholar]
- 8.Card JP, Moore RY. The suprachiasmatic nucleus of the golden hamster: immunohistochemical analysis of cell and fiber distribution. Neuroscience. 1984;13:415–431. doi: 10.1016/0306-4522(84)90240-9. [DOI] [PubMed] [Google Scholar]
- 9.Miyazato M, et al. Identification and functional analysis of a novel ligand for G protein-coupled receptor, Neuromedin S. Regul Pept. 2008;145:37–41. doi: 10.1016/j.regpep.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 10.Mori K, et al. Identification of neuromedin S and its possible role in the mammalian circadian oscillator system. Embo J. 2005;24:325–335. doi: 10.1038/sj.emboj.7600526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leak RK, et al. Suprachiasmatic pacemaker organization analyzed by viral transynaptic transport. Brain Res. 1999;819:23–32. doi: 10.1016/s0006-8993(98)01317-1. [DOI] [PubMed] [Google Scholar]
- 12.Morin LP. SCN organization reconsidered. J Biol Rhythms. 2007;22:3–13. doi: 10.1177/0748730406296749. [DOI] [PubMed] [Google Scholar]
- 13.Morin LP, et al. Complex organization of mouse and rat suprachiasmatic nucleus. Neuroscience. 2006;137:1285–1297. doi: 10.1016/j.neuroscience.2005.10.030. [DOI] [PubMed] [Google Scholar]
- 14.Evans JA, et al. Intrinsic regulation of spatiotemporal organization within the suprachiasmatic nucleus. PLoS One. 2011 doi: 10.1371/journal.pone.0015869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet. 2006;15(Spec No 2):R271–277. doi: 10.1093/hmg/ddl207. [DOI] [PubMed] [Google Scholar]
- 16.Lowrey PL, Takahashi JS. Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annu Rev Genomics Hum Genet. 2004;5:407–441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Busino L, et al. SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science. 2007;316:900–904. doi: 10.1126/science.1141194. [DOI] [PubMed] [Google Scholar]
- 18.Eide EJ, et al. Control of mammalian circadian rhythm by CKIepsilon-regulated proteasome-mediated PER2 degradation. Mol Cell Biol. 2005;25:2795–2807. doi: 10.1128/MCB.25.7.2795-2807.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Godinho SI, et al. The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science. 2007;316:897–900. doi: 10.1126/science.1141138. [DOI] [PubMed] [Google Scholar]
- 20.Shirogane T, et al. SCFbeta-TRCP controls clock-dependent transcription via casein kinase 1-dependent degradation of the mammalian period-1 (Per1) protein. J Biol Chem. 2005;280:26863–26872. doi: 10.1074/jbc.M502862200. [DOI] [PubMed] [Google Scholar]
- 21.Siepka SM, et al. Circadian mutant Overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell. 2007;129:1011–1023. doi: 10.1016/j.cell.2007.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gallego M, et al. An opposite role for tau in circadian rhythms revealed by mathematical modeling. Proc Natl Acad Sci U S A. 2006;103:10618–10623. doi: 10.1073/pnas.0604511103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu Y, et al. Modeling of a human circadian mutation yields insights into clock regulation by PER2. Cell. 2007;128:59–70. doi: 10.1016/j.cell.2006.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu Y, et al. Functional consequences of a CKIdelta mutation causing familial advanced sleep phase syndrome. Nature. 2005;434:640–644. doi: 10.1038/nature03453. [DOI] [PubMed] [Google Scholar]
- 25.Hirota T, et al. High-throughput chemical screen identifies a novel potent modulator of cellular circadian rhythms and reveals CKIalpha as a clock regulatory kinase. PLoS Biol. 2010 doi: 10.1371/journal.pbio.1000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meng QJ, et al. Setting clock speed in mammals: the CK1 epsilon tau mutation in mice accelerates circadian pacemakers by selectively destabilizing PERIOD proteins. Neuron. 2008;58:78–88. doi: 10.1016/j.neuron.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pittendrigh CS. Circadian rhythms and the circadian organization of living systems. Cold Spring Harb Symp Quant Biol. 1960;25:159–184. doi: 10.1101/sqb.1960.025.01.015. [DOI] [PubMed] [Google Scholar]
- 28.Pittendrigh CS, Daan S. Functional-Analysis of Circadian Pacemakers in Nocturnal Rodents. 5. Pacemaker Structure: A Clock for All Seasons. J Comp Physiol. 1976;106:333–355. [Google Scholar]
- 29.de la Iglesia HO, et al. Antiphase oscillation of the left and right suprachiasmatic nuclei. Science. 2000;290:799–801. doi: 10.1126/science.290.5492.799. [DOI] [PubMed] [Google Scholar]
- 30.Mason R. The effects of continuous light exposure on Syrian hamster suprachiasmatic (SCN) neuronal discharge activity in vitro. Neurosci Lett. 1991;123:160–163. doi: 10.1016/0304-3940(91)90920-o. [DOI] [PubMed] [Google Scholar]
- 31.Aschoff J. Exogenous and endogenous components in circadian rhythms. Cold Spring Harb Symp Quant Biol. 1960;25:11–28. doi: 10.1101/sqb.1960.025.01.004. [DOI] [PubMed] [Google Scholar]
- 32.Daan S, Pittendrigh CS. Functional-Analysis of Circadian Pacemakers in Nocturnal Rodents .3. Heavy-Water and Constant Light - Homeostasis of Frequency. J Comp Physiol. 1976;106:267–290. [Google Scholar]
- 33.Sudo M, et al. Constant light housing attenuates circadian rhythms of mPer2 mRNA and mPER2 protein expression in the suprachiasmatic nucleus of mice. Neuroscience. 2003;121:493–499. doi: 10.1016/s0306-4522(03)00457-3. [DOI] [PubMed] [Google Scholar]
- 34.Ohta H, et al. Constant light desynchronizes mammalian clock neurons. Nat Neurosci. 2005;8:267–269. doi: 10.1038/nn1395. [DOI] [PubMed] [Google Scholar]
- 35.Nagano M, et al. An abrupt shift in the day/night cycle causes desynchrony in the mammalian circadian center. J Neurosci. 2003;23:6141–6151. doi: 10.1523/JNEUROSCI.23-14-06141.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakamura W, et al. Differential response of Period 1 expression within the suprachiasmatic nucleus. J Neurosci. 2005;25:5481–5487. doi: 10.1523/JNEUROSCI.0889-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamazaki S, et al. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000;288:682–685. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]
- 38.Hazlerigg DG, et al. Photoperiod differentially regulates gene expression rhythms in the rostral and caudal SCN. Curr Biol. 2005;15:R449–450. doi: 10.1016/j.cub.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 39.Inagaki N, et al. Separate oscillating cell groups in mouse suprachiasmatic nucleus couple photoperiodically to the onset and end of daily activity. Proc Natl Acad Sci U S A. 2007;104:7664–7669. doi: 10.1073/pnas.0607713104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Naito E, et al. Reorganization of the suprachiasmatic nucleus coding for day length. J Biol Rhythms. 2008;23:140–149. doi: 10.1177/0748730408314572. [DOI] [PubMed] [Google Scholar]
- 41.Sosniyenko S, et al. Influence of photoperiod duration and light-dark transitions on entrainment of Per1 and Per2 gene and protein expression in subdivisions of the mouse suprachiasmatic nucleus. Eur J Neurosci. 2009;30:1802–1814. doi: 10.1111/j.1460-9568.2009.06945.x. [DOI] [PubMed] [Google Scholar]
- 42.Schaap J, et al. Heterogeneity of rhythmic suprachiasmatic nucleus neurons: Implications for circadian waveform and photoperiodic encoding. Proc Natl Acad Sci U S A. 2003;100:15994–15999. doi: 10.1073/pnas.2436298100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.VanderLeest HT, et al. Seasonal encoding by the circadian pacemaker of the SCN. Curr Biol. 2007;17:468–473. doi: 10.1016/j.cub.2007.01.048. [DOI] [PubMed] [Google Scholar]
- 44.Brown TM, Piggins HD. Spatiotemporal heterogeneity in the electrical activity of suprachiasmatic nuclei neurons and their response to photoperiod. J Biol Rhythms. 2009;24:44–54. doi: 10.1177/0748730408327918. [DOI] [PubMed] [Google Scholar]
- 45.Cambras T, et al. Circadian desynchronization of core body temperature and sleep stages in the rat. Proc Natl Acad Sci U S A. 2007;104:7634–7639. doi: 10.1073/pnas.0702424104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de la Iglesia HO, et al. Forced desynchronization of dual circadian oscillators within the rat suprachiasmatic nucleus. Curr Biol. 2004;14:796–800. doi: 10.1016/j.cub.2004.04.034. [DOI] [PubMed] [Google Scholar]
- 47.Lee ML, et al. Circadian timing of REM sleep is coupled to an oscillator within the dorsomedial suprachiasmatic nucleus. Curr Biol. 2009;19:848–852. doi: 10.1016/j.cub.2009.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schwartz MD, et al. Phase misalignment between suprachiasmatic neuronal oscillators impairs photic behavioral phase shifts but not photic induction of gene expression. J Neurosci. 2010;30:13150–13156. doi: 10.1523/JNEUROSCI.1853-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schwartz MD, et al. Dissociation of circadian and light inhibition of melatonin release through forced desynchronization in the rat. Proc Natl Acad Sci U S A. 2009;106:17540–17545. doi: 10.1073/pnas.0906382106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamaguchi S, et al. Synchronization of cellular clocks in the suprachiasmatic nucleus. Science. 2003;302:1408–1412. doi: 10.1126/science.1089287. [DOI] [PubMed] [Google Scholar]
- 51.Yan L, Okamura H. Gradients in the circadian expression of Per1 and Per2 genes in the rat suprachiasmatic nucleus. Eur J Neurosci. 2002;15:1153–1162. doi: 10.1046/j.1460-9568.2002.01955.x. [DOI] [PubMed] [Google Scholar]
- 52.Hamada T, et al. Temporal and spatial expression patterns of canonical clock genes and clock-controlled genes in the suprachiasmatic nucleus. Eur J Neurosci. 2004;19:1741–1748. doi: 10.1111/j.1460-9568.2004.03275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Foley NC, et al. Characterization of orderly spatiotemporal patterns of clock gene activation in mammalian suprachiasmatic nucleus. Eur J Neurosci. 2011 doi: 10.1111/j.1460-9568.2011.07682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yan L, et al. Exploring spatiotemporal organization of SCN circuits. Cold Spring Harb Symp Quant Biol. 2007;72:527–541. doi: 10.1101/sqb.2007.72.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Herzog ED, et al. Temporal precision in the mammalian circadian system: a reliable clock from less reliable neurons. J Biol Rhythms. 2004;19:35–46. doi: 10.1177/0748730403260776. [DOI] [PubMed] [Google Scholar]
- 56.Honma S, et al. Circadian periods of single suprachiasmatic neurons in rats. Neurosci Lett. 1998;250:157–160. doi: 10.1016/s0304-3940(98)00464-9. [DOI] [PubMed] [Google Scholar]
- 57.Herzog ED, et al. Clock controls circadian period in isolated suprachiasmatic nucleus neurons. Nat Neurosci. 1998;1:708–713. doi: 10.1038/3708. [DOI] [PubMed] [Google Scholar]
- 58.Aton SJ, et al. Vasoactive intestinal polypeptide mediates circadian rhythmicity and synchrony in mammalian clock neurons. Nat Neurosci. 2005;8:476–483. doi: 10.1038/nn1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu C, et al. Cellular construction of a circadian clock: period determination in the suprachiasmatic nuclei. Cell. 1997;91:855–860. doi: 10.1016/s0092-8674(00)80473-0. [DOI] [PubMed] [Google Scholar]
- 60.Low-Zeddies SS, Takahashi JS. Chimera analysis of the Clock mutation in mice shows that complex cellular integration determines circadian behavior. Cell. 2001;105:25–42. doi: 10.1016/s0092-8674(01)00294-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vogelbaum MA, Menaker M. Temporal chimeras produced by hypothalamic transplants. J Neurosci. 1992;12:3619–3627. doi: 10.1523/JNEUROSCI.12-09-03619.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nakamura W, et al. Clock mutation lengthens the circadian period without damping rhythms in individual SCN neurons. Nat Neurosci. 2002;5:399–400. doi: 10.1038/nn843. [DOI] [PubMed] [Google Scholar]
- 63.Underwood H, Calaban M. Pineal melatonin rhythms in the lizard Anolis carolinensis: I. Response to light and temperature cycles. J Biol Rhythms. 1987;2:179–193. doi: 10.1177/074873048700200302. [DOI] [PubMed] [Google Scholar]
- 64.Yoshida T, et al. Nonparametric entrainment of the in vitro circadian phosphorylation rhythm of cyanobacterial KaiC by temperature cycle. Proc Natl Acad Sci U S A. 2009;106:1648–1653. doi: 10.1073/pnas.0806741106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zimmerman WF, et al. Temperature compensation of the circadian oscillation in drosophila pseudoobscura and its entrainment by temperature cycles. J Insect Physiol. 1968;14:669–684. doi: 10.1016/0022-1910(68)90226-6. [DOI] [PubMed] [Google Scholar]
- 66.Francis AJ, Coleman GJ. The effect of ambient temperature cycles upon circadian running and drinking activity in male and female laboratory rats. Physiol Behav. 1988;43:471–477. doi: 10.1016/0031-9384(88)90121-7. [DOI] [PubMed] [Google Scholar]
- 67.Rajaratnam SM, Redman JR. Entrainment of activity rhythms to temperature cycles in diurnal palm squirrels. Physiol Behav. 1998;63:271–277. doi: 10.1016/s0031-9384(97)00440-x. [DOI] [PubMed] [Google Scholar]
- 68.Refinetti R. Entrainment of circadian rhythm by ambient temperature cycles in mice. J Biol Rhythms. 2010;25:247–256. doi: 10.1177/0748730410372074. [DOI] [PubMed] [Google Scholar]
- 69.Tokura H, Aschoff J. Effects of temperature on the circadian rhythm of pigtailed macaques Macaca nemestrina. Am J Physiol. 1983;245:R800–804. doi: 10.1152/ajpregu.1983.245.6.R800. [DOI] [PubMed] [Google Scholar]
- 70.Abraham U, et al. Coupling governs entrainment range of circadian clocks. Mol Syst Biol. 2010 doi: 10.1038/msb.2010.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brown SA, et al. Rhythms of mammalian body temperature can sustain peripheral circadian clocks. Curr Biol. 2002;12:1574–1583. doi: 10.1016/s0960-9822(02)01145-4. [DOI] [PubMed] [Google Scholar]
- 72.Buhr ED, et al. Temperature as a universal resetting cue for mammalian circadian oscillators. Science. 2010;330:379–385. doi: 10.1126/science.1195262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yoo SH, et al. PERIOD2∷LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ruby NF, et al. Circadian rhythms in the suprachiasmatic nucleus are temperature-compensated and phase-shifted by heat pulses in vitro. J Neurosci. 1999;19:8630–8636. doi: 10.1523/JNEUROSCI.19-19-08630.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Herzog ED, Huckfeldt RM. Circadian entrainment to temperature, but not light, in the isolated suprachiasmatic nucleus. J Neurophysiol. 2003;90:763–770. doi: 10.1152/jn.00129.2003. [DOI] [PubMed] [Google Scholar]
- 76.Vitaterna MH, et al. The mouse Clock mutation reduces circadian pacemaker amplitude and enhances efficacy of resetting stimuli and phase-response curve amplitude. Proc Natl Acad Sci U S A. 2006;103:9327–9332. doi: 10.1073/pnas.0603601103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bernard S, et al. Synchronization-induced rhythmicity of circadian oscillators in the suprachiasmatic nucleus. PLoS Comput Biol. 2007 doi: 10.1371/journal.pcbi.0030068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Webb AB, et al. Intrinsic, nondeterministic circadian rhythm generation in identified mammalian neurons. Proc Natl Acad Sci U S A. 2009;106:16493–16498. doi: 10.1073/pnas.0902768106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ko CH, et al. Emergence of noise-induced oscillations in the central circadian pacemaker. PLoS Biol. 2010 doi: 10.1371/journal.pbio.1000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Colwell CS. Rhythmic coupling among cells in the suprachiasmatic nucleus. J Neurobiol. 2000;43:379–388. doi: 10.1002/1097-4695(20000615)43:4<379::aid-neu6>3.0.co;2-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Long MA, et al. Electrical synapses coordinate activity in the suprachiasmatic nucleus. Nat Neurosci. 2005;8:61–66. doi: 10.1038/nn1361. [DOI] [PubMed] [Google Scholar]
- 82.Maywood ES, et al. Minireview: The circadian clockwork of the suprachiasmatic nuclei--analysis of a cellular oscillator that drives endocrine rhythms. Endocrinology. 2007;148:5624–5634. doi: 10.1210/en.2007-0660. [DOI] [PubMed] [Google Scholar]
- 83.Vosko AM, et al. Vasoactive intestinal peptide and the mammalian circadian system. Gen Comp Endocrinol. 2007;152:165–175. doi: 10.1016/j.ygcen.2007.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hughes AT, et al. Aberrant gating of photic input to the suprachiasmatic circadian pacemaker of mice lacking the VPAC2 receptor. J Neurosci. 2004;24:3522–3526. doi: 10.1523/JNEUROSCI.5345-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Watanabe K, et al. In vitro entrainment of the circadian rhythm of vasopressin-releasing cells in suprachiasmatic nucleus by vasoactive intestinal polypeptide. Brain Res. 2000;877:361–366. doi: 10.1016/s0006-8993(00)02724-4. [DOI] [PubMed] [Google Scholar]
- 86.An S, et al. Vasoactive intestinal polypeptide requires parallel changes in adenylate cyclase and phospholipase C to entrain circadian rhythms to a predictable phase. J Neurophysiol. 2011 doi: 10.1152/jn.00966.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Piggins HD, et al. Neuropeptides phase shift the mammalian circadian pacemaker. J Neurosci. 1995;15:5612–5622. doi: 10.1523/JNEUROSCI.15-08-05612.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Colwell CS, et al. Disrupted circadian rhythms in VIP- and PHI-deficient mice. Am J Physiol Regul Integr Comp Physiol. 2003;285:R939–949. doi: 10.1152/ajpregu.00200.2003. [DOI] [PubMed] [Google Scholar]
- 89.Harmar AJ, et al. The VPAC(2) receptor is essential for circadian function in the mouse suprachiasmatic nuclei. Cell. 2002;109:497–508. doi: 10.1016/s0092-8674(02)00736-5. [DOI] [PubMed] [Google Scholar]
- 90.Brown TM, et al. Disrupted neuronal activity rhythms in the suprachiasmatic nuclei of vasoactive intestinal polypeptide-deficient mice. J Neurophysiol. 2007;97:2553–2558. doi: 10.1152/jn.01206.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hughes AT, et al. Live imaging of altered period1 expression in the suprachiasmatic nuclei of Vipr2-/- mice. J Neurochem. 2008;106:1646–1657. doi: 10.1111/j.1471-4159.2008.05520.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Maywood ES, et al. Synchronization and maintenance of timekeeping in suprachiasmatic circadian clock cells by neuropeptidergic signaling. Curr Biol. 2006;16:599–605. doi: 10.1016/j.cub.2006.02.023. [DOI] [PubMed] [Google Scholar]
- 93.Ciarleglio CM, et al. Population encoding by circadian clock neurons organizes circadian behavior. J Neurosci. 2009;29:1670–1676. doi: 10.1523/JNEUROSCI.3801-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Albers HE, et al. Analysis of the phase shifting effects of gastrin releasing peptide when microinjected into the suprachiasmatic region. Neurosci Lett. 1995;191:63–66. doi: 10.1016/0304-3940(95)11559-1. [DOI] [PubMed] [Google Scholar]
- 95.Liu C, Reppert SM. GABA synchronizes clock cells within the suprachiasmatic circadian clock. Neuron. 2000;25:123–128. doi: 10.1016/s0896-6273(00)80876-4. [DOI] [PubMed] [Google Scholar]
- 96.McArthur AJ, et al. Gastrin-releasing peptide phase-shifts suprachiasmatic nuclei neuronal rhythms in vitro. J Neurosci. 2000;20:5496–5502. doi: 10.1523/JNEUROSCI.20-14-05496.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Smith RD, et al. Central administration of muscimol phase-shifts the mammalian circadian clock. J Comp Physiol A. 1989;164:805–814. doi: 10.1007/BF00616752. [DOI] [PubMed] [Google Scholar]
- 98.Aton SJ, et al. GABA and Gi/o differentially control circadian rhythms and synchrony in clock neurons. Proc Natl Acad Sci U S A. 2006;103:19188–19193. doi: 10.1073/pnas.0607466103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Travnickova-Bendova Z, et al. Bimodal regulation of mPeriod promoters by CREB-dependent signaling and CLOCK/BMAL1 activity. Proc Natl Acad Sci U S A. 2002;99:7728–7733. doi: 10.1073/pnas.102075599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.O’Neill JS, et al. cAMP-dependent signaling as a core component of the mammalian circadian pacemaker. Science. 2008;320:949–953. doi: 10.1126/science.1152506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ginty DD, et al. Regulation of CREB phosphorylation in the suprachiasmatic nucleus by light and a circadian clock. Science. 1993;260:238–241. doi: 10.1126/science.8097062. [DOI] [PubMed] [Google Scholar]
- 102.Tischkau SA, et al. Ca2+/cAMP response element-binding protein (CREB)-dependent activation of Per1 is required for light-induced signaling in the suprachiasmatic nucleus circadian clock. J Biol Chem. 2003;278:718–723. doi: 10.1074/jbc.M209241200. [DOI] [PubMed] [Google Scholar]
- 103.Lee B, et al. CREB influences timing and entrainment of the SCN circadian clock. J Biol Rhythms. 2010;25:410–420. doi: 10.1177/0748730410381229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Harmar AJ. Family-B G-protein-coupled receptors. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-12-reviews3013. REVIEWS3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rea MA. VIP-stimulated cyclic AMP accumulation in the suprachiasmatic hypothalamus. Brain Res Bull. 1990;25:843–847. doi: 10.1016/0361-9230(90)90179-4. [DOI] [PubMed] [Google Scholar]
- 106.Irwin RP, Allen CN. Neuropeptide-mediated calcium signaling in the suprachiasmatic nucleus network. Eur J Neurosci. 2010;32:1497–1506. doi: 10.1111/j.1460-9568.2010.07411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nielsen HS, et al. Vasoactive intestinal polypeptide induces per1 and per2 gene expression in the rat suprachiasmatic nucleus late at night. Eur J Neurosci. 2002;15:570–574. doi: 10.1046/j.0953-816x.2001.01882.x. [DOI] [PubMed] [Google Scholar]
- 108.Meredith AL, et al. BK calcium-activated potassium channels regulate circadian behavioral rhythms and pacemaker output. Nat Neurosci. 2006;9:1041–1049. doi: 10.1038/nn1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kent J, Meredith AL. BK channels regulate spontaneous action potential rhythmicity in the suprachiasmatic nucleus. PLoS One. 2008 doi: 10.1371/journal.pone.0003884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yamada Y, Forger D. Multiscale complexity in the mammalian circadian clock. Curr Opin Genet Dev. 2010;20:626–633. doi: 10.1016/j.gde.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Stoleru D, et al. Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature. 2004;431:862–868. doi: 10.1038/nature02926. [DOI] [PubMed] [Google Scholar]
- 112.Stoleru D, et al. A resetting signal between Drosophila pacemakers synchronizes morning and evening activity. Nature. 2005;438:238–242. doi: 10.1038/nature04192. [DOI] [PubMed] [Google Scholar]
- 113.Hong HK, et al. Inducible and reversible Clock gene expression in brain using the tTA system for the study of circadian behavior. PLoS Genet. 2007 doi: 10.1371/journal.pgen.0030033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Earnest DJ, Sladek CD. Circadian rhythms of vasopressin release from individual rat suprachiasmatic explants in vitro. Brain Res. 1986;382:129–133. doi: 10.1016/0006-8993(86)90119-8. [DOI] [PubMed] [Google Scholar]
- 115.Shinohara K, et al. Two distinct oscillators in the rat suprachiasmatic nucleus in vitro. Proc Natl Acad Sci U S A. 1995;92:7396–7400. doi: 10.1073/pnas.92.16.7396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kallo I, et al. Transgenic approach reveals expression of the VPAC2 receptor in phenotypically defined neurons in the mouse suprachiasmatic nucleus and in its efferent target sites. Eur J Neurosci. 2004;19:2201–2211. doi: 10.1111/j.0953-816X.2004.03335.x. [DOI] [PubMed] [Google Scholar]


