Abstract
The translocator protein (18 kDa; TSPO) is a mitochondrial drug- and cholesterol-binding protein that has been implicated in several processes, including steroidogenesis, cell proliferation, and apoptosis. Expression of the human TSPO gene is elevated in several cancers. To understand the molecular mechanisms that regulate TSPO expression in human breast cancer cells, the TSPO promoter was identified, cloned, and functionally characterized in poor-in-TSPO hormone-dependent, non-aggressive MCF-7 cells and rich-in-TSPO hormone-independent, aggressive, and metastatic MDA-MB-231 breast cancer cells. RNA ligase-mediated 5′-rapid amplification of cDNA ends analysis indicated transcription initiated at multiple sites downstream of a GC-rich promoter that lacks functional TATA and CCAAT boxes. Deletion analysis indicated that the region from −121 to +66, which contains five putative regulatory sites known as GC boxes, was sufficient to induce reporter activity up to 24-fold in MCF-7 and nearly 120-fold in MDA-MB-231 cells. Electrophoretic mobility shift and chromatin immunoprecipitation assays indicated that Sp1, Sp3 and Sp4 bind to these GC boxes in vitro and to the endogenous TSPO promoter. Silencing of Sp1, Sp3 and Sp4 gene expression reduced TSPO levels. In addition, TSPO expression was epigenetically regulated at one or more of the identified GC boxes. Disruption of the sequence downstream of the main start site of TSPO differentially regulated TSPO promoter activity in MCF-7 and MDA-MB-231 cells, indicating that essential elements contribute to its differential expression in these cells. Taken together, these experiments constitute the first in-depth functional analysis of the human TSPO gene promoter and its transcriptional regulation.
Keywords: TSPO, breast cancer, metastasis, Sp1, Sp3, epigenetics
1. INTRODUCTION
The translocator protein (18 kDa) (TSPO), previously known as the peripheral-type benzodiazepine receptor, is a widely expressed 18-kDa mitochondrial protein that is biochemically and pharmacologically distinct from the central benzodiazepine receptor binding sites that allosterically regulate the GABAA receptor [1–3]. Subcellular fractionation and radioligand binding studies have shown that TSPO is predominantly localized to the mitochondria [4] and is concentrated at contact sites between the outer and inner mitochondrial membranes [5]. TSPO associates with the voltage dependent anion channel and adenine nucleotide translocator [6–8], which together contribute to the formation of the mitochondrial permeability transition pore [9]. The functional role of TSPO has been best described in steroidogenic cells, in which it acts as a high affinity cholesterol-binding protein that participates in the intra-mitochondrial transport of cholesterol, the rate-determining step in the synthesis of steroids [10]. Additional functions have been suggested by the pharmacological effects of high affinity TSPO ligands, which have been shown to modulate mitochondrial respiration, cell proliferation, apoptosis, and differentiation, although the mechanisms underlying these effects are poorly understood.
The levels of TSPO expression vary according to tissue and cell type and can be altered pathologically. In normal tissues, high levels of TSPO expression are observed in the adrenal cortex, steroidogenic cells of the gonads, and clusters of differentiated cells within glandular epithelia. Increased expression of the human TSPO gene has been described in several cancers, including high-grade glioblastomas, prostate, ovarian, colon, and breast carcinomas [11]. In breast cancer cell lines and clinical specimens, expression of TSPO mRNA and radioligand binding and/or immunoreactivity increases in a manner that correlates positively with invasiveness and/or malignancy [12–14]. The mechanism by which TSPO gene expression is increased physiologically in specific cell types and pathologically in tumors is poorly understood. We have previously reported the presence of TSPO gene amplification in aggressive metastastic breast cancer cells and biopsies [11,15]. However, TSPO gene amplification does not appear to be sufficient to account for the levels of increased expression of TSPO in aggressive human breast cancer cells without contributions from additional mechanisms of aberrant gene expression. Thus, we investigated the mechanism(s) regulating transcription of the TSPO gene in MCF-7 and MDA-MB-231 cells, which express relatively low and high levels, respectively, of TSPO mRNA and protein [14].
In the present study, we aimed at to functionally characterize the human TSPO promoter and to investigate its transcriptional regulation in breast cancer cells, as well as the similarities in its structure and regulation to that of the mouse promoter. As an initial step in the analysis of transcriptional regulation, the promoter directing expression of the human TSPO gene in breast cancer cell lines was identified, cloned, sequenced, and functionally characterized to determine the minimal sequence necessary to support basal levels of promoter activity. Specific substitution mutations were then introduced into the proximal region identified as necessary for maintaining near-maximal promoter activity, in order to define important regulatory elements. Based on the functional analysis of the TSPO promoter, potential protein/DNA interactions were investigated using electrophoretic mobility shift assay (EMSA) and supershift analyses. In silico analysis of the cloned human TSPO promoter sequence revealed high GC content in the proximal region of the promoter, whereas further analysis showed that the TSPO gene is situated within a CpG island. A methylation inhibitor (5-Azacytidine) and deacetylation inhibitor (TSA) were used to reveal the involvement of epigenetic mechanisms, such as methylation and acetylation, in the regulation of TSPO gene expression. As a result, this work constitutes the first functional description of the promoter of the human TSPO gene and compares and contrasts its regulation in two breast cancer cell lines that can be distinguished on the basis of hormonal dependence, chemotactic and chemoinvasive potential, and differential expression of several markers of malignancy, including differential expression of the TSPO gene.
2. Materials and methods
2.1. Cell culture
MDA-MB-231 and MCF-7 human breast cancer cell lines were obtained from the Cell Culture Core Facility of Lombardi Comprehensive Cancer Center (LCCC) and maintained in Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum. (GIBCO, Burlington, ON). Human mammary epithelial cells (HMECs) from Cambrex, Inc. (East Rutherford NJ) were maintained in Mammary Epithelial Growth Medium (MEGM; Clonetics-Lonza, Walkersville, MD). Drosophila SL2 cells (D.Mel.2) were purchased from Invitrogen (Carlsbad, CA) and maintained as previously described [16]. Both MCF-7 and MDA-MB-231 cell lines were subjected to DNA fingerprinting and tested for mycoplasma contamination as part of routine maintenance by LCCC.
2.2. RNA ligase-mediated 5′-rapid amplification of cDNA ends (RLM-5′RACE)
Total RNA was isolated using TRIzol Reagent, according to the manufacturer’s instructions (Invitrogen). RLM-5′RACE was performed using the GeneRacer Kit, according to the manufacturer’s instructions (Invitrogen). 5′RACE-ready cDNA libraries were prepared by reverse transcription using SuperScript II RT and total RNA isolated from HMEC, MCF-7, and MDA-MB-231 cells. Commercial RACE-ready cDNA libraries prepared from normal human testes and kidney were purchased from Invitrogen. Nested PCR was performed using a commercial nested 5′ primer in combination with a reverse gene-specific primer complementary to either TSPO exon 3 or exon 4. Amplified PCR products were gel-purified and subcloned into pCR4-TOPO (Invitrogen) for sequencing. Primers used in 5′-RACE are listed in Supplemental Table S1.
2.3. Plasmid construction
The putative promoter sequence was amplified by PCR using the GC-Rich PCR System (Roche Molecular Biochemicals, Indianapolis, IN) and BAC clone dJ526I14 (Wellcome Trust Sanger Institute, Cambridge, UK; GenBank accession number z82214) as template. Amplified TSPO promoter fragments were directionally cloned into the promoter-less pGL3-Basic reporter vector (Promega, Madison, WI) using Mlu I and Bgl II restriction sites incorporated into the forward and reverse PCR primers, respectively. Promoter sequences were verified using Taq FS Dye Terminator Cycle Sequencing performed by the LCCC DNA Sequencing Core Facility. Primers used in the construction of deletion and substitution mutants are listed in Supplemental Table S1. Substitution mutants were prepared by PCR using either a splicing by overlap extension or conventional ligation protocol to join promoter fragments containing 6-bp substitutions, creating Xba I, Pst I, or Nhe I restriction sites in place of the wild-type sequence. All potential substitution mutants were evaluated by a search of the MatInspector v2.2 database [17] to ensure that the mutated sequence did not create a known consensus DNA binding motif. In the splicing by overlap extension protocol, two sets of primer pairs were used in the initial PCR to generate products that overlapped at the position where the mutation was to be inserted. In the second PCR, a mutated double-stranded TSPO promoter fragment was amplified from the overlapping primary PCR products using flanking primers hybridizing to positions −121 and +66 of the TSPO promoter.
2.4. Transient transfections and luciferase reporter assays
All cells were transfected in 12-well plates using Lipofectamine Plus reagent (Invitrogen). Each transfection consisted of 150 fmol of the specific TSPO promoter-pGL3Basic reporter construct, 10 ng of pRLTK (Promega), and the necessary amount of pUC19 plasmid to bring the total amount of plasmid DNA per transfection to 845 ng. In epigenetic analyses, MDA-MB-231 and MCF-7 cells were treated with 50 nM Trichostatin A (TSA) or 5 μM 5-Azacytidine (AZA) 24 hrs after transfection with the F11 −121/+66 promoter construct and lysates were harvested 24 hrs later. The luciferase activities of all lysates were measured using the Dual Luciferase Reporter Assay System (Promega) and a Wallac Victor2 equipped with two reagent injectors (Perkin Elmer, Waltham, MA).
2.5. Quantitative real time (QRT)-PCR
MDA-MB-231 and MCF-7 cells were seeded in 6-well plates and cell lysates were collected 24 h later. RNA was extracted using the RNeasy kit (Qiagen, Valencia, CA), according to the manufacturer’s instructions. RNA (1 μg) was reverse transcribed using the TaqMan reverse transcription reagents kit (Roche) in a 20-μl final volume, according to manufacturer’s instructions. The resulting cDNA was diluted to 60 μl final volume with RNase free water and used as a template for real-time PCR using TSPO specific primers and SYBR GREEN dye. Amplification of 18S ribosomal RNA served as a control.
2.6. Immunoblotting
MDA-MB-231 and MCF-7 cells were seeded at 150,000 cells per well in 6-well plates for 24 h. In the epigenetic analyses, cells were treated with either TSA or 5-AZA for 24 h before harvesting and extracting protein. Cells were lysed in 1X cold RIPA buffer (Cell Signaling Technology, Danvers, MA) supplemented with Halt protease inhibitor cocktail 1:100 (Pierce, Rockford, IL). Protein concentration was measured using a standard Bradford assay (Bio-Rad, Hercules, CA), and equal amounts of protein were electrophoresed on 4–20% tris-glycine polyacrylamide gels (Invitrogen), transferred to PVDF of nitrocellulose membranes, and probed with antibodies to Sp1 and Sp3 (1:500, Santa Cruz Biotechnology), TSPO (1:1000) [3], GAPDH (1:2000) (Trevigen, Gaithersburg, MD), and HPRT (1:1000, Abcam, Cambridge, MA). Nuclear lysates (20 μg) from MCF-7 and MDA-MB-231 were separated by polyacrylamide gel electrophoresis, transferred to nitrocellulose membranes, and probed with antibodies for Sp1, Sp3 and Sp4 (1:500, Santa Cruz Biotechnology, Santa Cruz, CA). Immunoreactive proteins were visualized by enhanced chemiluminescence (Amersham, Piscataway, NJ) using a FUJI ImageQuant image reader LAS4000 (GE Healthcare, Piscataway, NJ) for capturing images or film processing. Densitometry of each band was measured using Multi-gauge V3.0 software.
2.7. Electromobility shift assay (EMSA)
Gel shifts were performed using the LightShift Chemiluminescent EMSA Kit (Perbio Science, Rockford, IL), according to an optimized adaptation of the manufacturer’s protocol. Nuclear extracts were prepared using the Nuclear Extract Kit from Active Motif (Carlsbad, CA). Binding reactions were incubated at room temperature for 20 min, electrophoresed on a 6% Tris-borate-EDTA non-denaturing polyacrylamide gel (Invitrogen), transferred to BrightStar-Plus nylon membranes (Ambion, Austin, TX), and visualized using streptavidin-horseradish peroxidase chemiluminescence. Competition analyses were performed using 100- to 200-fold excess of the unlabeled oligonucleotide duplex. Supershift analyses were performed after a 2-h pre-incubation on ice of the reaction components with anti-Sp1 (PEP2X, Santa Cruz Biotechnology) and/or anti-Sp3 (D-20X, ibid.) or anti-Sp4 (Santa Cruz Biotechnology) prior to the addition of the biotinylated probe. Sequences of oligonucleotide probes used for EMSA are included in Supplemental Table S2.
2.8. Chromatin immunoprecipitation (ChIP)
ChIP was performed using the Magna ChIP G kit (Millipore, Lake Placid, NY), according to the manufacturer’s protocol. Briefly, MDA-MB-231 and MCF-7 cells were grown in 150-mm dishes to 80% confluence and then cross-linked with medium containing 1% formaldehyde for 10 min at room temperature. Plates were washed three times with ice cold 1X PBS before being scraped and pelleted by centrifugation (800 × g) for 5 min at 4°C. Cells were lysed in buffer (provided by Millipore) containing protease inhibitors and nuclei were collected by centrifugation (800 × g) for 5 min at 4°C. Nuclei were suspended in nuclear lysis buffer (provided by Millipore) containing protease inhibitors and kept on ice. Nuclear DNA was sheared to an average size of 200–1000 bp with eight 10-sec pulses at 25% power using a Vibracell VC 130 sonicator (Sonics and Materials Inc., Newtown, CT) fitted with a 3-mm stepped microtip. After shearing, chromatin was immunoprecipitated with 2 μl Sp1, Sp3 or Sp4 (Santa Cruz Biotechnology) antibodies overnight at 4°C with rotation, and complexes were captured using magnetic protein-G beads. Magnetic beads were washed with a low and high salt immune complex wash buffer, LiCl immune complex wash buffer, and then TE buffer, following the manufacturer’s instructions. Cross-links were reversed by incubating beads in a rotating oven for 4 h at 62°C. DNA was purified and diluted 1:10 before being subjected to QRT-PCR using primers specific for the human TSPO proximal promoter (forward 5′-AGCTCACTCCTCAAAGCCACCTCTC-3′ and reverse 5′-AGGAGGAGGAGCCGAGAGACTTGCT -3′). Immunoprecipitates obtained with normal 2 μl rabbit IgG (#2729s, Cell Signaling Technology) served as negative controls. For ChIP after TSA treatment cells were grown in 150-mm dishes to 70% confluence before being treated with DMSO or TSA for an additional 24 h, after which cells were cross-linked and subjected to the procedure described above.
2.9. Sp1, Sp3 and Sp4 siRNA treatments
MDA-MB-231 and MCF-7 cells were seeded at 150,000 cells per well in 6-well plates. Cells were transfected 24 h later with 2 μg of the Sp1, Sp3 and Sp4 siRNA pools or 2 μg of each for Sp1/Sp3 combined transfections (Santa Cruz, Biotechnology), using Lipofectamine 2000 (Invitrogen), as recommended by the manufacturer. For the Sp1, Sp3 and Sp4 siRNA pools, MDA-MB-231 and MCF-7 cells were cultured for 72 h before being harvested for immunoblotting. Target gene knockdown was verified by QRT-PCR (data not shown) and immunoblotting. TSPO knockdown was verified by immunoblot analysis of cell extracts, when indicated. To measure the effect of Sp1 and Sp3 siRNA knockdown on promoter activity, cells were transfected with siRNA for 24 h before transfecting the F11 plasmid construct, as described above, for an additional 24 h before measuring luciferase activity.
2.10. Bisulfite sequencing
The CpG islands searcher software was used to identify CpG islands in the vicinity of the TSPO promoter [18]. To isolate genomic DNA for bisulfite sequencing, cells were washed with PBS, collected and snap frozen in liquid nitrogen. Genomic DNA was extracted using the DNeasy kit (Qiagen), according to the manufacturer’s instructions, and digested with EcoRI and NotI. After purification,, a DNA sample (2 μg) was bisulfite-treated using the EpiTect Bisulfite kit (Qiagen) and eluted in a 20-μl volume. The PCR mix for the nested PCR amplification of the bisulfite treated DNA consisted of 0.2 mM dNTP, 1.5 mM MgCl2, 0.2 μM primers, 1U PlatinumTaq polymerase (Invitrogen), and 2 μl bisulfite-treated DNA in a 30-μl total volume. The conditions for the first PCR reaction were: pre-incubation at 94°C (5 min), 30 cycles of 30 sec 94°C denaturation, 30 sec annealing at 50°C, 3 min extension at 72°C, and 4 min incubation at 72°C. The nested PCR was carried out with 1 μl of the first PCR product under the following conditions: pre-incubation at 94°C (3 min), 30 cycles of 30-sec 94°C denaturation, 30 sec at 52°C, 30-min extension at 72°C, and 4-min incubation at 72°C. Primer sequences for the first PCR reaction were hTSPO5S - ATTATGGGGAGGAGGAGGAGT and hTSPO5A – ACCTCAATTTCCCTCTATCC, and for the second PCR reaction, were hTSPO5S and hTSPO3A - AAAACACTACCTAACAAAAATTAACCA. After PCR, 30 μl PCR product was mixed with loading dye and run on a 2% agarose gel containing ethidium bromide. The 253-bp bands were cut, gel purified, and ligated into the pGEM-T easy vector (Promega) for sequencing. CpG viewer software was used to graphically represent the bisulfite sequencing results [19].
2.11. Sequence and statistical analysis
Sequence analysis was performed using Vector NTI software (Informax, Invitrogen). Statistical analysis was performed using Prism version 4.0 (GraphPad Software, San Diego, CA). Group means were compared using the Student’s t test or two-way analysis of variance (ANOVA). Data are presented as mean ± standard error of the mean (SEM), where p< 0.05 was considered significant.
3. Results
3.1. Identification of transcription start site(s) (tss)
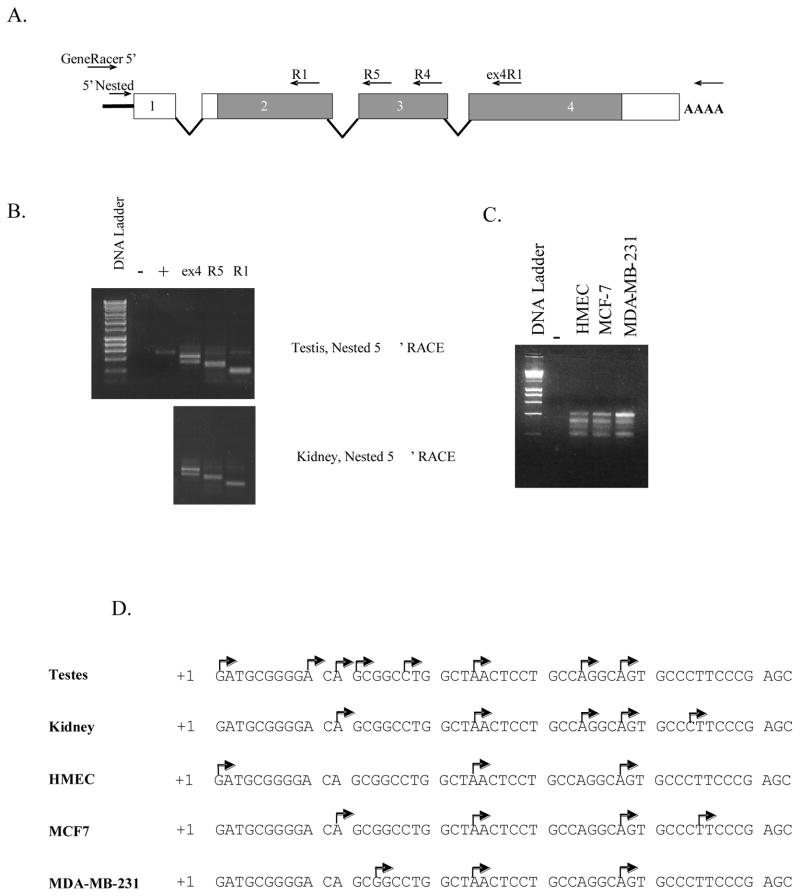
To identify the promoter used to direct transcription of the human TSPO gene in a variety of cellular contexts, RLM 5′ RACE-ready cDNA libraries were prepared from HMEC, MDA-MB-231, and MCF-7 cell lines and normal human testes and kidneys. An antisense gene-specific primer complementary to sequences in TSPO exon 4 and nested primers complementary to sequences in either exon 4 or exon 3 were used for two successive rounds of PCR (Figure 1A). The number of visible bands amplified in the RLM-5′RACE nested PCR depended on the source of the RNA and the gene-specific primer used (Figure 1B and 1C). Sequence analysis of 10 or more clones per library, selected on the basis of insert size, indicated that the TSPO gene is expressed using a common promoter in all tissues and cell lines examined, with alternative splicing of TSPO mRNA accounting for the multiplicity of PCR products observed using specific primer pairs (Supplemental S3). The identities of all alternative transcripts were confirmed by sequencing RT-PCR products that were generated using primers that anneal complementary to the cloning vector or to the TSPO 5′ and 3′ UTR (data not shown; primers listed in Supplemental Table S1).
Figure 1.
Mapping of transcription initiation sites using RLM-5′RACE. (A) Schematic representation of primers used for primary and nested PCR, and their relative positions complementary to sequences of the fourth, third, and second exons of the TSPO gene. (B) Nested PCR amplification products obtained from reactions using different gene-specific primers and RACE-ready cDNA libraries prepared from human testis and kidney as the initial template. (C) Nested PCR amplification products in which RACE-ready cDNA libraries prepared from total RNA isolated from HMEC, MCF-7, or MDA-MB-231 cell lines were used as the initial template and primer ex4R1 was used as gene-specific primer. Plus and minus signs denote positive and negative PCR controls, respectively. (D) Map of the transcription initiation window used by the TSPO gene in the tissue or cell line shown. Each bent arrow represents a transcription start site identified by sequence analysis of subcloned nested PCR products.
Multiple transcription start sites (tss) were mapped to an approximate 40 bp window for each cell- and tissue-type. The the first nucleotide of the clone mapping to the most upstream position from the human testes cDNA library was arbitrarily designated as +1 (Figure 1D) and a total of eight different start sites mapping to a region extending from +1 to +38 were observed.. Similarly, five different start sites were mapped to a region extending from +12 to +44 using a kidney cDNA library. A mixture of common and unique initiation sites was observed for HMEC, MCF-7, and MDA-MB-231 cells, although the distribution of these sites was less diverse than the tss distribution observed using libraries prepared from whole tissue. In every cell line and tissue examined, two common sites were observed at nucleotide positions +24 and +38. Sequence analysis suggested that the relative frequency of initiation at the common sites may vary between tissues and cell lines (data not shown). These results show that a common promoter is used to initiate transcription of the TSPO gene at multiple sites within a window of 40–50 bases in a variety of cellular and tissues, including normal human mammary epithelial cells and breast cancer cell lines with differing invasive and chemotactic properties.
3.2. Functional characterization of the TSPO promoter: TSPO gene expression is higher in MDA-MB-231 cells than in MCF-7 cells
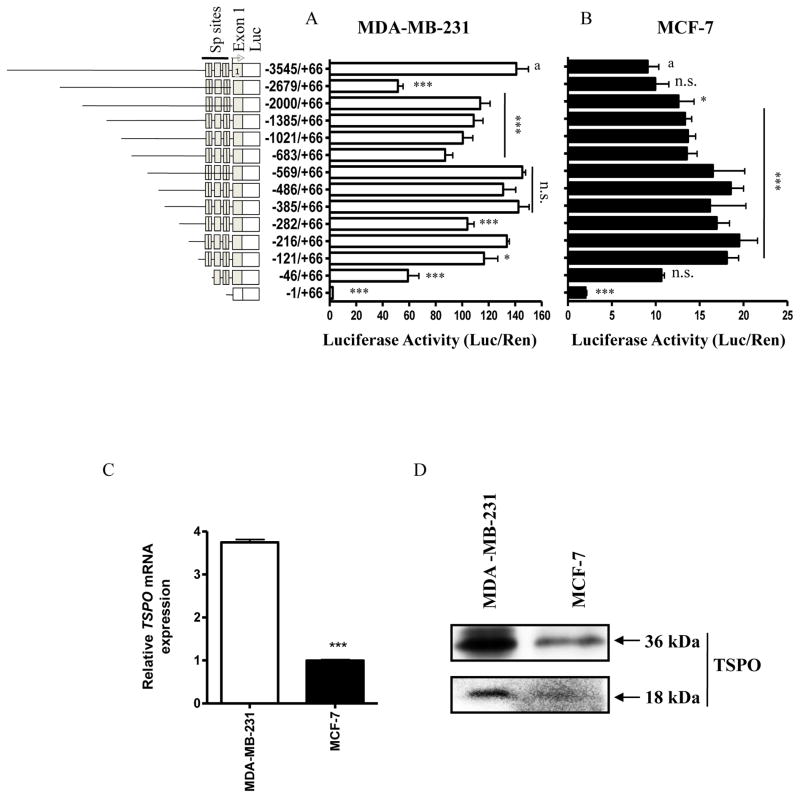
After identifying the TSPO promoter, we next asked i) whether the adjacent 5′ flanking region is sufficient to direct TSPO promoter activity in these cell lines; and ii) whether regulatory elements within this flanking region may contribute to the differing levels of TSPO gene expression exhibited by these cells [14]. A set of fourteen 5′ deletion mutants was constructed by amplifying putative promoter sequences by PCR from BAC clone dJ526I14 and subcloning these sequences into the pGL3-Basic vector. Basal promoter activity of the full length (−3545/+66) promoter was substantially higher in MDA-MB-231 cells (roughly 140-fold over background) compared to MCF-7 cells (12-fold). Deletion analysis in MDA-MB-231 cells indicated the presence of a strongly activating regulatory element between −3545 and −2679 which did not appear to contribute to TSPO promoter activity in MCF-7 cells (Figure 2A and B). Additional deletions showed that near maximal promoter activity could be obtained in both cell types using a construct with as few as 121 bases of flanking sequence. Subsequent deletion to nucleotide −46 dramatically reduced promoter activity in each cell type, ranging from a 35% reduction in MCF-7 cells to over 50% reduction in MDA-MB-231 cells. The addition deletion of 45 bases was sufficient to reduce promoter activity to levels minimally greater than background. Similar analyses in MCF-7 cells with different passage histories (P<35 vs. P>150), which are sometimes used as a model of cancer progression, did not reveal any significant differences except for slightly higher promoter activity overall (data not shown) in the higher passage cells. Similar analysis of deletion mutants in HepG2 cells also indicated that near-maximal promoter activity could be obtained with the proximal −121/+66 construct (data not shown); thus, the proximal TSPO promoter appears to be sufficient to reconstitute near-maximal promoter activity in a variety of cellular contexts, although additional distal elements may be active in MDA-MB-231 and HepG2 cells that are necessary to overcome the effects of distal inhibitory elements. Since TSPO promoter activity was significantly higher in MDA-MB-231 cells compared to MCF-7 cells (around 8-fold higher), we checked the level of TSPO mRNA (Figure 2C) and protein (Figure 2D) in both cells and found both the 18-kDa TSPO monomer and 36-kDa TSPO dimer levels to be significantly higher in the former compared to the latter, which agrees with the promoter activity.
Figure 2.
Functional characterization of the TSPO promoter. A series of plasmids containing 5′ unidirectional deletions of the TSPO promoter was prepared by PCR, subcloned into pGL3Basic, and transfected in equimolar amounts, along with 10 ng pRLTK, into MCF-7 and MDA-MB-231 cells. Luciferase activity was measured 48 hr post-transfection using a dual luciferase assay. The obtained luciferase activities are shown as fold activation over background obtained with the promoter-less vector pGL3 Basic. Data are presented as the mean of three or more experiments performed in triplicate, and error bars represent one standard deviation. (A) Pooled results of four independent experiments in which MDA-MD-231 and (B) MCF-7 cells were transiently transfected with 5′ unidirectional deletions of the TSPO promoter. (C) Relative TSPO mRNA levels in MDA-MB-231 vs. MCF-7 cells. (D) Immunoblot analysis of TSPO protein levels in MDA-MB-231 vs. MCF-7 cells. All constructs evaluated in Figure 2A and B demonstrated significant luciferase activity (p< 0.05) compared to the promoter-less pGL3-Basic control, except promoter construct −1/+66 in which all GC boxes had been deleted. *p < 0.05, **p < 0.01, ***p < 0.001 vs full promoter (a), n.s., non-significant.
Overall, these experiments demonstrated that i) the flanking region upstream of the transcription initiation window is sufficient to activate promoter activity in a manner that correlates with TSPO mRNA levels in both MCF-7 and MDA-MB-231 cells; ii) a distal positively acting element may be necessary to achieve maximal activity in MDA-MB-231 cells; and iii) near maximal promoter activity can be reconstituted in each cell line with as little as 121 bases of flanking sequence.
3.3. GC box 3 and overlapping GC boxes 4 and 5 must be intact for full basal TSPO promoter activity in MDA-MB-231 and MCF-7 cell lines
Sequence and database analysis of the human TSPO promoter revealed that no TATA box or consensus CCAAT boxes are found in the vicinity of the transcription initiation window. The GrailEXP database indicated that the TSPO promoter is situated within a CpG island (67% C+G, 0.73 Obs./Exp) that extends approximately 470 bp upstream and 615 bp downstream of the transcription initiation window [20]. Within the proximal promoter, potential binding sites for several transcription factors were observed, including AP2, Ets/FliI, EGR1, MZF1, MAZ, and multiple binding motifs, known as GC boxes, for members of the Specificity Protein/Krüppel-like factor (Sp/KLF) family of transcription factors (Figure 3A). When organized on the basis of core binding motifs (GGGCGG), the −121/+66 construct, which we have designated as the TSPO proximal promoter, contained five potential GC boxes, two of which partially overlapped proximally (GC.1/2) and two of which partially overlapped distally (GC.4/5). An additional canonical GC box (GC.3) was found to be situated at a position between the overlapping GC.1/2 and GC.4/5 boxes. The linear organization of these potential Sp1-like binding sites differs from the murine Tspo proximal promoter, which we previously showed to consist of two overlapping Sp1-like binding sites flanked on each side by canonical GC boxes.
Figure 3.
Conserved GC boxes between mouse and human TSPO promoters are essential for promoter activity. (A) Alignment of the proximal promoters of the human and mouse TSPO genes, emphasizing the relative positions of GC boxes (denoted GC.1–5 on the human promoter and Sp.1–4 on the mouse promoter) on each promoter. The position and orientation of core binding motifs are shown by horizontal arrows. Bent arrows above the line denote the most upstream start site (designated +1) and the two common transcription start sites observed for the human promoter. The bent arrow below the aligned sequence denotes the most commonly observed start site for the mouse TSPO promoter (designated +1). The beginning of intron 1 of the TSPO gene is shown in lowercase letters. (B) Site-directed mutagenesis of putative GC boxes in the proximal TSPO promoter. A series of luciferase reporter plasmids with mutations targeting putative binding motifs GC.1, GC.2, GC.3, GC.4, and GC.5 was constructed in the context of the −121/+66 promoter construct. Luciferase activity that was significantly different (p < 0.05) from that observed with the wild-type promoter construct (−121/+66) is indicated by an asterisk.
To define the boundaries and functional status of potential regulatory elements within the proximal promoter, we performed sequential site-directed substitution mutations throughout the region flanking the transcription initiation window. Within this upstream region, the mutations targeting putative GC boxes resulted in the greatest reduction in promoter activity (Fig. 3B). Mutation of GC box 3 (GC3) resulted in substantial losses of promoter activity, decreasing by as much as 60% in MCF-7 cells and 50% in MDA-MB-231 cells. Mutation of the overlapping GC boxes 4 and 5 also decreased promoter activity by 20–35% in each cell line. In contrast, targeted mutation of GC boxes 1 and 2 did not decrease levels of basal promoter activity in either cell line. Mutation of 5′ flanking sequence in a region not containing a known binding motif (−76 to −71) did not reduce promoter activity. Together, these results indicate that GC box 3 and overlapping GC boxes 4 and 5 must be intact for full basal promoter activity in breast cancer cell lines.
3.4. Specific binding of transcription factors Sp1 and Sp3 to the proximal TSPO promoter in vitro
The binding of Sp1 and Sp3 transcription factors to GC boxes has been shown to be involved in both the constitutive activation of housekeeping genes and the regulation of tissue-specific and inducible genes. To determine whether the putative GC boxes within the proximal TSPO promoter are sufficient to specifically bind these transcription factors, EMSAs were performed using probes designed to include one or more of the GC boxes. Expression of Sp1 and Sp3 proteins was verified in MCF-7 and MDA-MB-231 cells by immunobloting of nuclear extracts harvested from these cell lines (data not shown). To evaluate potential protein-DNA interactions, EMSAs were performed using probes designed to include one or more of the putative GC boxes of the proximal TSPO promoter. When equal amounts of nuclear extracts prepared from sub-confluent MCF-7 and MDA-MB-231 cells were incubated with biotinylated probes, at least two protein-DNA complexes were observed (Figures 4A and B). The specificity of this interaction was demonstrated by competition using 100- to 200-fold excess of unlabeled, wild-type probe (Figures 4A and B, lanes 3 or 4). In contrast, unlabeled probes containing 2-bp mutations targeting putative Sp1 binding sites did not compete for binding (Figures 4A and B, lanes 4 or 3). Pre-incubation of extracts with antibodies to either Sp1 or Sp3 resulted in either the altered mobility or elimination of specific complexes. Pre-incubation with anti-Sp1 antibody resulted in a partial supershift of the upper complex, designated complex 1 (Figures 4A and B, lane 7), while pre-incubation with anti-Sp3 resulted in the complete disruption of the lower complex, designated complex 2, and the partial supershift of the complex 1 (Figures 4A and B, lane 8). Similar results were obtained using oligonucleotides specific for either GC box 3 (Figure 4B) or overlapping GC boxes 4/5 and 1/2 (Figure 4A). A residual complex of higher mobility which occurred on a subset of EMSAs, designated complx3, was not altered by incubation with either anti-Sp1 or anti-Sp3 (Figures 4A and B, band3). To investigate whether this could be due to the presence of Sp4 binding, we performed supershift analyses using probes corresponding to either GC Box4/5 or GC Box 3 and nuclear extracts from MDA-MB-231 cells. Incubation with an antibody to Sp4 did not result in either the formation of an observable complex of slower mobility or a decrease in the amount of retarded complex formed compared to the reaction in which probe is incubated with nuclear extract alone (Figure 4C).
Figure 4.
GC Box motifs from the proximal TSPO promoter can bind to both Sp1 and Sp3 proteins, in vitro. (A) EMSA and supershift analyses comparing interactions of proteins in nuclear extracts isolated from MCF-7 and MDA-MB-231 cells with probes corresponding to either overlapping GC Boxes 1/2 or GC Boxes 4/5. (B) EMSA and supershift analysis using a probe corresponding to GC Box 3. For competition experiments, unlabeled wild-type (wt), double-stranded core Sp1-like binding motif mutants (mu), and unrelated DNA sequence [Epstein Barr virus nuclear antigen binding site (E)] were used at 100-fold molar excess. (C) EMSA and supershift analyses comparing interactions of proteins in nuclear extracts isolated from MDA-MB-231 and MCF-7 cells with probes corresponding to GC Box 3.. For competition experiments, unlabeled wt and double-stranded core Sp1-like binding motif mu were used at 100-fold molar excess. Specific complexes in all EMSAs are designated 1–3, where “1” denotes a specific retarded complex that is minimally comprised of overlapping complexes formed by Sp1 and/or Sp3 binding to the respective probe; “2” denotes a specific retarded complex formed by the binding of an Sp3 isoform to the respective probe; and “3” denotes a specific complex of unknown composition.
3.5. Sp1, Sp3, and Sp4 bind the endogenous TSPO promoter
The relative expression of Sp1, Sp3 and Sp4 isoforms was assessed in MDA-MB-231 and MCF-7 cells (Figure 5A). Multiple studies have previously looked at the differential expression of Sp isoforms in a panel of breast cancer cell lines [21,22]. We next used chromatin immunoprecipitation (ChIP) assays to determine whether Sp1, Sp3 and Sp4 transcription factors are bound to the endogenous TSPO promoter in intact cells. Data shown in Figure 5B demonstrate that both Sp1 and Sp3 were bound to the endogenous TSPO promoter in both MDA-MB-231 and MCF-7 cells, whereas the IgG control reaction produced negative results. To further validate the ability of Sp4 to bind the TSPO endogenous promoter in intact MDA-MB-231 cells, we performed ChIP using Sp4 antibody. Figure 5C shows that Sp4 did bind endogenous TSPO promoter in both MDA-MB-231 and MCF-7 cells, whereas the IgG control reaction produced negative results.
Figure 5.
Sp1, Sp3, and Sp4 are bound to the endogenous TSPO promoter in intact MDA-MB-231 and MCF-7 cells. (A) Relative expression levels of Sp1, Sp3 and Sp4 isoforms in MDA-MB-231 and MCF-7 cells. (B) DNA from MDA-MB-231 and MCF-7 cells was precipitated with antibodies specific for Sp1 and Sp3 in ChIP assays. (C) DNA from MDA-MB-231 and MCF-7 cells was precipitated with antibodies specific for Sp4 and Sp3 in ChIP assays. DNA was amplified by QRT-PCR using primers specific for the proximal area of the TSPO promoter. Normal rabbit IgG served as a negative control. Data were analyzed as percent of input and fold enrichment compared to IgG, and are derived from three independent experiments (n = 9). **p < 0.01, ***p < 0.001 vs. control IgG.
3.6. Activation of the TSPO promoter by Sp1 and Sp3
To test whether Sp1 and/or Sp3 activates transcription of the TSPO promoter, transient transfection experiments were performed using Drosophila SL2 cells, which lack detectable activation of GC boxes by members of the Sp family of transcription factors. Expression plasmids for Sp1 or Sp3 (pPacSp1, pPacSp3, respectively) were co-transfected with a reporter plasmid containing either the wild-type −121/+66 promoter sequence or specific GC box mutants. Cell lysates were harvested 24 h after transfection and results were reported as fold-activation relative to the parental vector, pPac0. Both Sp1 and Sp3 expression plasmids activated the wild-type construct in a dose-dependent manner, albeit to only moderate levels (Figure 6A). When expressed in combination, the expression plasmids had an additive effect on promoter activity (Figure 6B). In contrast, overexpressing Sp1 and Sp3 in breast cancer cells had contrasting effects on TSPO proximal promoter activity. Overexpressing Sp1 slightly induced TSPO promoter activity in MDA-MB-231 cells at higher doses (Figure 6C). Sp3 had variable effects, dependent on the dose in MDA-MB-231 cells (Figure 6C); however, both Sp1 and Sp3 reduced TSPO promoter activity in MCF-7 cells (Figure 6D). Together, these results suggest that additional mechanisms may regulate Sp1 and Sp3 function in controlling TSPO promoter activity in breast cancer cells that are not constitutively present in SL2 cells.
Figure 6.
Activation of the human TSPO promoter by Sp1 and Sp3 in Drosophila SL2 cells. Sp1/Sp3 differentially regulates the TSPO promoter in MDA-MB-231 and MCF-7 cells. Drosophila SL2 cells were co-transfected with equimolar amounts of wild-type pGL3(−121/+66) or related GC box mutant plasmids along with expression plasmids for Sp1 (pPacSp1) or Sp3 (pPacSp3). Results are plotted as fold activation over background, as determined by co-transfection of the empty expression vector pPac0. Data are reported as the mean of four independent experiments performed in quadruplicate and error bars represent one standard deviation. (A) Transient transfection of SL2 cells with increasing amounts of pPacSp1 or pPacSp3. (B) Co-transfection of SL2 cells with a constant amount of pPacSp1 (25 ng) and increasing amounts of pPacSp3 (0–25 ng). (C) Co-transfection of MDA-MB-231 cells with increasing amounts of pPacSp1 or pPacSp3. (D) Transient transfection of MCF-7 cells with increasing amounts of pPacSp1 or pPacSp3. (*p < 0.05, **p < 0.01, ***p < 0.001, n.s., non-significant) denotes luciferase activity that is significantly different from that observed with the wild-type promoter (−121/+66) marked as (a).
3.7. Sp1 and Sp3 siRNA significantly reduce TSPO expression levels in MDA-MB-231 and MCF-7 cells
To confirm the role of Sp1 and Sp3 transcription factors in the regulation of TSPO expression, pools of Sp1 and Sp3 siRNA were used to lower Sp1 and Sp3 mRNA and protein levels, respectively, in MDA-MB-231 and MCF-7 cells. Figures 7A and B show that Sp1 and Sp3 siRNA pools were each able to reduce TSPO promoter activity in these cell lines, an effect that was further enhanced when Sp1 and Sp3 siRNAs were combined. At the protein level, combined Sp1/Sp3 knockdown was necessary to achieve the most significant reduction in TSPO protein levels (Figures 7C and F), as shown by densitometry of the blots in Figures 7D and 7G. These results demonstrate that siRNA-mediated targeting of Sp1 and Sp3 differentially reduced TSPO expression in MDA-MB-231 and MCF-7 cells (Figures 7C, D, F and G), suggesting that these transcription factors may contribute to the overall levels of TSPO expression in these cells by different mechanisms. The effectiveness of siRNA knowckodown of Sp1 and Sp3 proteins was confirmed by immunoblot analysis in Figures 7E and H.
Figure 7.
Sp1 and Sp3 regulate TSPO expression in MDA-MB-231 and MCF-7 cells. (A and B) Sp1 specific siRNA significantly reduced TSPO promoter activity, in both MDA-MB-231 and MCF-7 cells (C and F), as well as reducing 18-kDa TSPO protein levels. Sp3 siRNA significantly reduced TSPO promoter activity, as well as 18-kDA TSPO protein levels in MCF-7 (B, F and G); however, it did not seem to reduce TSPO protein levels alone (F and G). On the other hand, (C, D, F and G) combining Sp1 and Sp3 siRNA synergistically reduced TSPO protein levels in MDA-MB-231 and MCF-7 cells to levels lower than that of Sp1 alone, implicating a combinatorial regulation of both isoforms in regulating TSPO expression. (D and G) Densitometry analysis of the blots in C and F. (E and H) immunoblots representing effectiveness of the siRNAs used to knockdown Sp1 and Sp3 proteins respectively in MDA-MB-231 and MCF-7 cells. Blots are representative of at least three experiments. *p < 0.05, **p < 0.01, ***p < 0.001.
3.8. Sp4 siRNA significantly reduces TSPO expression levels in MDA-MB-231 and MCF-7 cells
Since Sp4 was able to bind to the endogenous TSPO promoter, we investigated the ability of a Sp4 siRNA pool to reduce TSPO expression. Indeed, knockdown of Sp4 protein levels (Figures 8C and F) significantly reduced TSPO expression in MDA-MB-231 (Figures 8A and B) and MCF-7 cells (Figure 8D and E), albeit with higher efficiency in the MCF-7 cells. Interestingly, Sp4 was shown by ChIP to bind the endogenous in the ChIP assay (Figure 5C).
Figure 8.
Sp4 regulates TSPO expression in MDA-MB-231 and MCF-7 cells. (A and D) Sp4 specific siRNA significantly reduced 18-kDa TSPO protein levels in both MDA-MB-231 and MCF-7 cells. (B and E) Densitometry analysis of the blots in A and D. (C and F) immunoblots representing effectiveness of siRNA used to knockdown the Sp4 protein in MDA-MB-231 and MCF-7 cells respectively. Blots are representative of at least three experiments. *p < 0.05, and ***p < 0.001.
3.9. TrichostatinA (TSA) can induce TSPO promoter activity in MDA-MB-231 and MCF-7 cells
We next investigated whether TSPO gene expression is modulated by epigenetic mechanisms in breast cancer cell lines. As discussed earlier, the TSPO promoter is situated within a CpG island that extends approximately 470 bp upstream and 615 bp downstream of the transcription initiation window [20]. Analysis of the promoter area between −183/+35, which encompasses the five GC boxes we studied above, revealed the presence of 25 CpG dinucleotides (Figure 9A). Using bisulfite modification to investigate the methylation status of these CpG dinucleotides, we found that the TSPO promoter was partially methylated in MCF-7 cells, but exhibited relatively little methylation in MDA-MB-231 cells (Figure 9B.1). Since TSPO is considered a housekeeping gene, it is not surprising that it is not highly methylated. To determine whether incomplete methylation made a functional difference on the TSPO promoter, cells transfected with the proximal promoter (−121/+66) construct were treated with the DNA methyltransferase inhibitor 5-azacytidine. Consistent with their respective methylation profiles, treatment with 5-AZA had little effect on TSPO promoter activity in MDA-MB-231 cells, although activity was induced roughly 3-fold following the treatment of MCF-7 cells (Figure 9D.2). In contrast, treating with the histone deacetylase inhibitor TSA strongly induced TSPO promoter activity in both cell lines (Figure 9C.2) and reduced the methylation signature on the TSPO promoter in MCF-7 cells. Interestingly, TSA induced TSPO promoter activity in MCF-7 cells at a level 10-fold higher than in MDA-MB-231 cells. In addition, TSA reduced the methylation status of some CpG islands in MCF-7 cells. To investigate the role played by methylation, we overexpressed the F11 TSPO promoter construct and treated cells with 5-Azacytidine (AZA). Treating with AZA reduced the methylation of some CpG islands in MCF-7 cells, yet it only induced promoter activity by 3-fold compared to 120-fold with TSA (Figure 9D.2). These results suggest that the interplay epigenetic modulations within the proximal TSPO promoter can dramatically alter promoter activity and may be critical to regulating levels of TSPO expression in different cell types.
Figure 9.
Methylation status of predicted CpG islands in proximal TSPO promoter. TSA but not AZA induced TSPO promoter activity. (A) Twenty-five CpG islands were identified in the proximal area of TSPO promoter area −183/+35, which encompasses the previously identified GC boxes, using searcher software. (B.1, C.1, D.1) Methylation status of predicted CpG islands in the TSPO promoter was studied using genomic DNA extracted from MDA-MB-231 and MCF-7 cells, treated or not with TSA or AZA for 24 h. Genomic DNA (1.5 μg) was bisulfate modified using EpiTect Bisulfite kit. (B.2) Cells were transiently transfected with the F11 construct of TSPO promoter and luciferase activity was measured 24 h later. Comparative levels of basal TSPO promoter activity in MDA-MD-231 and MCF-7 cells. (C.2) Twenty-four hours after the transient transfection, cells were treated with TSA for 24 h before measuring luciferase activity. (D.2) Twenty-four hours after the transient transfection, cells were treated with AZA for 24 h before measuring luciferase activity. Results are derived from three independent experiments (n = 9) **p < 0.01, ***p < 0.001 vs. control.
In addition, it is apparent that TSA reduced the methylation status of multiple CpGs. In the case of the endogenous promoter, it is possible that TSA may induce TSPO promoter activity by preventing deacetylation of the histones, reducing methylation and leaving the promoter open to transcription factors that bind and induce its activity.
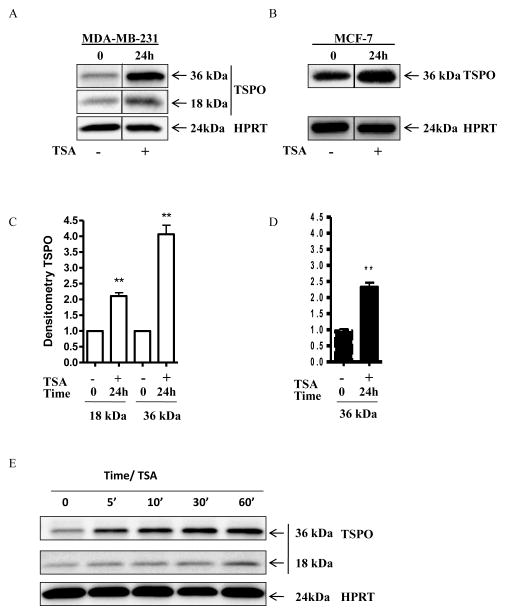
3.10. TSA induced TSPO protein levels at longer and shorter time points
MDA-MB-231 and MCF-7 cells were treated with TSA for 24 h and TSPO protein levels were analyzed by immunoblotting. TSA induced the levels of both 18-kDa and 36-kDa TSPO in MDA-MB-231 cells (Figure 10A and C), as well as the levels of the 36-kDa TSPO in MCF-7 cells (Figure 10B and D). In contrast to MDA-MB-231 cells, the levels of 18-kDa TSPO in MCF-7 cells were low to undetectable (Figure 2D). Interestingly, treating MDA-MB-231 cells with TSA for shorter time points led to a rapid increase in the 36-kDa TSPO dimer, whereas only a slight increase in the intensity of the 18-kDa band was observed (Figure 10E).
Figure 10.
TSA induced TSPO expression and TSPO oligomerization. MDA-MB-231 and MCF-7 cells were treated with TSA for 24 h before protein lysates were analyzed by immunoblotting for TSPO expression levels. (A) TSA induced 18-kDa and 36-kDa TSPO expression 24 h after treatment of MDA-MB-231 cells. (B) Levels of 18-kDa TSPO were too low to be detectable in MCF-7 cells even after TSA treatment; however, 36-kDa TSPO was significantly induced. (C, D) Densitometric analysis of the blots. (E) TSA treatment for the indicated time points showed rapid increases of the 36-kDa TSPO dimer.
3.11. GC3 is essential for basal and TSA induced TSPO promoter activity
TSA activation of TSPO promoter activity appears to be mediated by elements in the −121/+66 region, since deletion of the distal region between −3545/−121 caused no significant reduction in the induced TSPO promoter activity. To determine whether TSA mediates its effect through one or more of the previously identified GC boxes, cells transfected with promoter constructs containing mutations in the respective GC boxes were treated with TSA and the effect on TSPO promoter activity was assessed. The canonical GC box, GC3, was required for mediating both basal (Figure 3B) and TSA-inducible promoter activity (Figure 11A). in MCF-7 and MDA-MB-231 cells (Figure 11A). Mutating GC.1/2, which did not affect basal promoter activity, had a modest but significant effect on TSA-inducibility in MDA-MB-231 cells, but not MCF-7.
Figure 11.
TSA induced TSPO promoter activity through box GC3, and Sp1 and Sp3 differentially alter TSA effect on TSPO promoter in MDA-MB-231 and MCF-7 cells.(A) A series of luciferase reporter plasmids with mutations targeting putative binding motifs GC.1, GC.2, GC.3, GC.4, and GC.5 was constructed in the context of the −121/+66 promoter construct, along with the full length promoter and a −76 mutation that does not target a known element, were transiently expressed in MDA-MB-231 and MCF-7 cells treated (black bars) or not (white bars) with TSA for 24 h. Luciferase activity was measured 24 h after TSA treatment. Empty vector was used as a control. Results are derived from three independent experiments (n = 9) **p < 0.01, ***p < 0.001 vs. the wild-type promoter. (B) Co-transfection of MDA-MB-231 cells with increasing amounts of pPacSp1 or pPacSp3 along with F11 construct and treating on not with TSA for 24 h before measuring luciferase activity. (C) Co-transfection of MCF-7 cells with increasing amounts of pPacSp1 or pPacSp3 along with F11 construct and treating on not with TSA for 24 h before measuring luciferase activity. Sp1, Sp3, and Sp4 reamain bound to the endogenous TSPO promoter in intact (D) MDA-MB-231 and (E) MCF-7 cells after 24 h treatment with TSA. DNA from MDA-MB-231 and MCF-7 cells treated with TSA was precipitated with antibodies specific for Sp1, Sp3, and Sp4 in ChIP assays. DNA was amplified by QRT-PCR using primers specific for the proximal area of the TSPO promoter. Normal rabbit IgG served as a negative control. Data were analyzed as percent of input and fold enrichment compared to IgG, and are derived from three independent experiments (n = 9). **p < 0.01, ***p < 0.001 vs. control IgG.
The effects of overexpressing Sp1 and Sp3 cells on TSA-inducibility of TSPO promoter activity was also investigated in MCF-7 and MDA-MB-231 cells. The ability of the TSPO promoter to be superactivated by TSA was elevated in MDA-MB-231 cells by transfection with a small amount of Sp1 expression plasmid, although intermediate amounts of plasmid did not appear to affect TSA-inducibility. TSA-inducibility in MDA-MB-21 cells was more responsive to titration with increasing amounts of Sp3 expression plasmid, which increased TSA-inducibility by as much as 65% (Figure11B). In contrast, increasing Sp1 or Sp3 levels in MCF-7 cells decreased TSA-inducibility in a dose-dependent manner (Figure11C). ChIP assays showed that Sp1 and Sp3 continued to bind to the endogenous TSPO promoter following TSA treatment, although the relative ratio of factors bound appears to change with treatment (Figure11D and E).
3.12. The promoter region downstream of the main transcription start site contributes to the higher TSPO promoter activity in MDA-MB-231 cells compared to MCF-7 cells
To examine whether a downstream control element contributes to the overall strength of the TSPO promoter in MCF-7 and MDA-MB-231 cells, 3′ deletion mutants were constructed in which sequences from +66 to +13 were removed. Deletion analysis indicated that the TSPO promoter in MCF-7 cells was only moderately affected by deleting of all exon 1 sequence, with the activity of the −121/+13 construct roughly equivalent to that of the −121/+66 construct, which contains all of exon 1 except for three nucleotides surrounding the splice junction (Figure 12B). Similarly, inclusion of exon 1 sequence was not required to maintain maximum TSPO promoter activity in HepG2 cells, a hepatocellular carcinoma cell line that expresses low to moderate levels of TSPO mRNA (data not shown). Thus, exon 1 sequence, which contributes to the 5′ UTR of TSPO mRNA, may not be necessary to obtain maximal promoter activity in cell lines that exhibit weak promoter activity and express low levels of TSPO protein.
Figure 12.
Promoter area downstream of the main transcription start site contributes to the higher TSPO promoter activity in MDA-MB-231 cells compared to MCF-7 cells. MDA-MB-231 and MCF-7 cells were transfected with a series of 3′ deletion constructs with different lengths of exon 1. (A) Deletion analysis indicated that the TSPO promoter in MCF-7 cells was only moderately affected by deletion of all exon 1 sequence, with the activity of the −121/+13 construct roughly equivalent to the −121/+66 construct, which contains most of exon 1. In contrast, at least two different regions of downstream sequence were necessary for full TSPO promoter activity in MDA-MB-231 cells. Inclusion of additional sequence increased promoter activity in a step-wise manner in MDA-MB-231 cells. MDA-MB-231 and MCF-7 cells were transiently transfected with a series of mutant constructs at the indicated bases, and luciferase activity was measured after 24 h. Mutation of the sequences at position +38 resulted in a more dramatic gain of activity by the TSPO promoter in MCF-7 (C) cells compared to MDA-MB-231 cells (B).
In contrast, at least two different regions in the downstream sequence were necessary for near maximal TSPO promoter activity in MDA-MB-231 cells (Figure 12A). One important region corresponded to the sequences immediately adjacent to the commonly used transcription initiation site at position +38. Inclusion of additional sequence increased promoter activity in a step-wise manner, suggesting that more than one regulatory element may be functional within this region. Similarly, the requirement of exon 1 sequence for maximal promoter activity was also observed in analysis of the 3′ deletions in MA-10 cells, a steroidogenic mouse Leydig tumor cell line which expresses high levels of TSPO mRNA and protein (data not shown). Similar to MDA-MB-231 cells, inclusion of the sequence immediately downstream of the common tss at +38 increased promoter activity by 2-fold, although MA-10 cells exhibited a slightly different requirement for the most downstream sequences of exon 1 to achieve maximum activity. Together, these results suggest that the region extending from +38 to +66 is an important determinant of TSPO promoter strength, which may be necessary to up-regulate TSPO promoter activity in cells that express higher levels of TSPO (Figure 12A).
To better define the boundaries of the regulatory elements contributing to the differential effect of downstream sequences on TSPO promoter activity, a panel of 6-bp substitution mutants was prepared and analyzed. Given the 6-fold difference in promoter activity exhibited by these cell lines, the resulting data were normalized to the activity exhibited by the wild-type promoter sequence for each cell line to facilitate comparisons between the cell lines. Interestingly, the effects of substitution mutagenesis were less dramatic than the deletion mutations. Mutation of the sequence between +44 and +61 resulted in diminished TSPO promoter activity in both cell lines (Figures 12B and C). In MDA-MB-231 cells, mutation of the sequence between +44 and +61 resulted in diminished promoter activity in three of the four constructs evaluated (Figure 12B), with the +50/55 construct exhibiting wild-type levels of activity. If this region represents the boundary between two regulatory elements, then the combined effects of the mutations at nucleotides +61 and +44 would be expected approach the loss of activity observed by deletion of the same region. Additional mutation of the sequence at position +38 resulted in a gain of TSPO activity in both cell lines, although this effect was considerably more dramatic in MCF-7 cells (Figures 12B and C). Database analysis of the area between +40/+66 revealed the presence of multiple potential transcription factor binding sites, including the signal transducer and activator of transcription 3 (STAT3) and v-ets erythroblastosis virus E26 oncogene homolog (Ets), as well as Aryl hydrocarbon receptor (AhR) and its dimerization partner, the AhR nuclear translocator (ARNT). These results suggest that the region including and immediately downstream of the transcription initiation window may be an additional determinant of promoter strength in cells that differentially express TSPO protein, although the mechanisms by which this downstream region contributes to transcriptional regulation remain to be investigated.
4. Discussion
Previously, we have shown that the TSPO gene is differentially expressed in a panel of human breast cancer cell lines and clinical specimens in a manner correlating negatively with estrogen receptor status and positively with increasing malignant phenotype [13,14]. In the present study, we address the hypothesis that differences in transcriptional regulation of the TSPO gene contribute to the differences in expression observed in these cell lines and tumor biopsies. To model the increasing malignant character of tumor biopsies, we performed functional analysis of the TSPO gene in MCF-7 cells, a more differentiated, less invasive, ER-positive breast cancer cell line that weakly expresses TSPO, and MDA-MB-231 cells, which are a less differentiated, more invasive, ER-negative breast cancer cell line that strongly expresses TSPO.
The work described in this manuscript provides the first in-depth description of the human TSPO promoter and its transcriptional regulation. Similar to the mouse Tspo promoter, 5′-RLM-RACE indicated that the human TSPO promoter is a TATA-less promoter situated within a CpG island in both MCF-7 and MDA-MB-231 cell lines [16]. The promoter of the human TSPO gene was shown to direct transcription from multiple sites within a 40- to 50-bp window in a variety of tissue and cellular contexts. Multiple transcription start sites within a loosely defined region are typically characteristic of TATA-less promoters located within CpG islands. Each cell line and tissue examined exhibited an array of common and unique transcription sites. Interestingly, two of these sites were used in all tissues and cell types examined.
To determine whether different regulatory elements mediate the differences in transcription of the TSPO gene observed in MCF-7 and MDA-MB-231 cells, we functionally analyzed the promoter identified by RLM-5′RACE in each cell line. Basal promoter activity of the full length (−3545/+66) promoter roughly correlated with the relative levels of TSPO expression by each cell line, with MDA-MB-231 cells showing up to 6-fold more promoter activity than MCF-7 cells. Deletion mutagenesis indicated the presence of a distal regulatory element between -3545 and -2679 which was strongly activating in MDA-MB-231 cells, but not MCF-7 cells, which may be characterized in a future investigation. For the remainder of this study, we investigated the regulatory mechanisms acting on the −121/+66 construct, designated the TSPO proximal promoter, which was sufficient to reconstitute near-maximal promoter activity in each cell line. Within this region, database analysis identified five putative GC boxes, regulatory elements which are commonly found in the promoters of genes which are ubiquitously expressed. These regulatory elements typically function as high affinity binding sites for Sp1, Sp3, Sp4 proteins, although additional GC-binding transcription factors have been reported.
Alignment of the TSPO promoter with the mouse Tspo proximal promoter revealed areas of conservation and divergence. In the human promoter, two of the GC boxes overlapped proximally (GC.1/2) and two overlapped distally (GC.4/5), with a single canonical GC box (GC.3) located at a position between the overlapping motifs. In contrast, the murine Tspo promoter was organized as central overlapping GC boxes (Sp1.2.3) flanked distally (Sp1.4) and proximally (Sp1.1) by canonical GC boxes. Additional areas of conservation in the two promoters included the regions surrounding the most distal tss (+1) observed human cells and tissues and the region including the common tss at nucleotide +38. The region of conservation upstream of the tss at the +1 nucleotide includes the overlapping GC boxes 1 and 2, which correspond to the Sp2 and Sp3 elements of the mouse promoter. The region corresponding to the Sp1 element of the murine promoter is also conserved; however, this sequence more closely resembles a MZF-1 motif in the human promoter due to the presence of an adenine at position +10. Additional conserved sequences are found in the vicinity of GC.4, although neither the overlapping core motif (GGGCGG) of GC.5 nor the single GC.3 motif is present in the mouse promoter.
Whether these interspecies differences in promoter architecture result in differences in the regulation of transcriptional is not known. Targeted mutation of GC boxes 1 and 2 did not decrease levels of promoter activity in either breast cancer cell line. In contrast, mutation of GC box 3 resulted in substantial loss of activity, with promoter activity decreasing by as much as 60% in MCF-7 cells and 50% in MDA-MB-231 cells. Mutation of GC boxes 4 and 5 decreased promoter activity by 20–35% in each cell line. Our previous analysis of the mouse Tspo promoter showed a similar dependence on the central motif, despite its overlapping core binding sequences, with minor contributions from the more distal canonical GC box (Sp1.4) [16]. Together, these results suggest that central and distal GC boxes within the proximal promoter must be intact for near-maximal basal activity in a variety of contexts, including human breast cancer cell lines and mouse steroidogenic cells.
EMSA and supershift experiments demonstrated that Sp1 and Sp3 from MDA-MB-231 and MCF-7 nuclear extracts bind to the isolated GC Box 3 and the overlapping motifs of GC Boxes 1/2 and GC Boxes 4/5 in vitro. In addition, mutations targeting the core motifs of these GC boxes both reduced proximal promoter activity of luciferase reporter constructs and eliminated competition by these elements in gel shift assays. It should be noted that in the EMSA and anti-Sp1 and Sp3 supershift experiments with both MDA-MB-231 and MCF-7 nuclear extracts, there was a residual retarded band that was not supershifted (Figures 4A and B, band 3). This could be due to the presence of Sp4 binding. Sp4 protein expression has been reported in several breast cancer cell lines [22], including the MCF-7 and MDA-MB-231 cell lines. We performed supershift analyses using probes corresponding to either GC Box4/5 or GC Box 3 and nuclear extracts from MDA-MB-231 cells. Incubation with an antibody to Sp4 did not result in either the formation of an observable complex of slower mobility nor a decrease in the amount of retarded complex formed, compared to the reaction in which probe was incubated with nuclear extract alone. Since the binding of Sp1, Sp3, and Sp4 to GC-rich oligos has previously been shown to form retarded complexes with overlapping mobilities, these results suggest that any contribution of Sp4 binding to the probes corresponding to GC boxes 4/5 and 3 in supershift experiments is likely to be minimal compared to the binding of Sp1 and Sp3. Alternatively, ChIP was employed to verify the ability of Sp1, Sp3 and Sp4 to bind the endogenous TSPO promoter and regulate its expression.
Although EMSA and ChIP analyses indicate that Sp1 and Sp3 bind to GC4/5, GC3, and GC1/2 in vitro and ChIP showed Sp4 binding to the endogenous TSPO proximal promoter in intact cells, it does not provide information regarding the function of bound proteins. Sp1 and Sp3 are both bifunctional, acting as either an activator or inhibitor of transcription depending on factors such as post-translational modification, isoform expression, and promoter architecture and context [23]. Expression studies in Drosophila SL2 cells, which are deficient in GC box-binding Sp/KLF family members, showed that expression of Sp1 or Sp3, either alone or in combination, is sufficient to activate the TSPO promoter, albeit at relatively low levels. The additive effect of Sp1 and Sp3 co-expression contrasts with some promoters, where Sp3 co-expression diminishes the ability of Sp1 to activate promoter activity [14, 24]. Since alterations in the relative levels of Sp1 and Sp3 expression have been shown to be important in regulating cell proliferation and tumor progression [25–27], we also investigated the effects of over-expressing Sp1 and Sp3 in breast cancer cells on TSPO proximal promoter activity. Titrating increasing amounts of Sp1 and Sp3 had little effect on promoter activity in MDA-MB-231 cells, except when the largest amount of pPacSp3 is used (50ng), although transfecting increasing amounts of either pPacSp1 or pPacSp3 was sufficient to repress proximal promoter activity in MCF-7 cells. Whether these results reflect actual competition for binding to the TSPO promoter, differential autoregulatory mechanisms, or off-target effects of these transcription factors is not known. Additionally, combined siRNA pools targeting Sp1, Sp3 and Sp4 were able to significantly reduce Sp1, Sp3 and Sp4 protein levels and TSPO expression in both MDA-MB-231 and MCF-7 cells. These results confirmed the role of Sp proteins on TSPO expression regulatory elements.
Since the mutation of GC boxes had proportional effects on TSPO promoter activity in both MCF-7 and MDA-MB-231 cells, we also examined the sequences including and downstream of the transcription initiation window to see if additional regulatory elements are present which influence promoter activity in these cells. Deletion and substitution analyses suggested that two or more activating regulatory elements are present between +39 and +66 which have a greater effect on TSPO promoter activity in MDA-MB-231 cells than MCF-7 cells, and an inhibitory element between +38/+43 which has greater influence on promoter activity in MCF-7 cells. Interestingly, these elements are located within the region of conservation observed in the alignment of the human and mouse promoters. Analysis of the human TSPO promoter 3′ deletion mutants in MA10 cells, which strongly express TSPO, showed a similar requirement for these downstream sequences for maximal promoter activity (data not shown). Together, these results suggest that sequence dependent mechanisms within the region surrounding and downstream of the +38 tss may be required for full promoter activity in cells that highly express TSPO. Whether these mechanisms include the enrichment of transcription initiation at +38 through Inr function or the action of regulatory elements that differentially modulate TSPO promoter activity in different cell types remains to be determined.
Epigenetic modification of chromatin and DNA may provide additional mechanisms by which promoter activity may regulated in MCF-7 and MDA-MB-231 cells. Treatment with 5-azacytidine showed that methylation appears to regulate TSPO promoter activity in MCF-7 cells, but not MDA-MB-231 cells. Partial methylation was observed in 4/11 sequences in the region separating GC.4/5 and GC.3, suggesting that steric interference at these binding sites may influence promoter activity. However, the methylation of more distal CpGs outside of the region considered in this study may also influence endogenous promoter activity, particularly CpG nucleotides that are immediately adjacent in intron 1.
In contrast to the effects of 5-azacytidine, which only induced promoter activity in MCF-7 cells, treatment with a histone deacetylase inhibitor (TSA) strongly activated the TSPO proximal promoter in both MCF-7 and MDA-MB-231 cells. Inhibiting histone deacetylase activity to increase histone acetylation provides a signature which can be read by other proteins and destabilizes higher chromatin order to provide greater access to transcription machinery. While this mechanism could account for the observed TSA-inducibility, histone acetyltransferases and histone deacetylases also have non-histone substrates and can modulate gene expression by directly acetylating or deacetylating transcription factors and cofactors. Sp1 and Sp3 have both been reported to be acetylated [28,29], although the functional implication of this post-translational modification is not clear. Post-translational modifications could alter the transactivating activities of these factors. Alternatively, synergistic interactions that are present in MDA-MB-231 cells, but absent in MCF-7 cells may control cell-specific mechanisms regulating TSPO gene expression. Sp1 has been reported to have synergistic interactions with numerous transcription factors [23]. Given the results of our 3′ deletion analysis, we hypothesize that interactions between a protein bound to the downstream region and Sp1 bound to one or more of the GC boxes synergistically activates transcription in cells which highly express TSPO, such as Leydig cells and MDA-MB-231 cells. Similarly, the absence of binding of this protein to the downstream region or the binding of a protein which does not synergistically interact with Sp1 may lock the TSPO promoter in a less active functional state, such as normal tissues that ubiquitously express low levels of TSPO or in well-differentiated, ER-positive breast cancer cell lines like MCF-7 cells.
Since Sp1, Sp3 and Sp4 did not seem to be the sole regulators of human TSPO expression, we investigated whether TSPO gene expression is modulated by epigenetic mechanisms. Interestingly, treating MDA-MB-231 cells with TSA for periods as short as 5 min resulted in the formation of the 36-kDa TSPO dimer, which increased in a time-dependent manner. Since this time is not sufficient for TSA to act at the transcriptional level, it remains to be determined how TSA induced TSPO dimer formation. This phenomenon may be due to TSA inducing TSPO mRNA stability or translation, or TSA-induced acetylation of TSPO at a lysine residue leading to TSPO dimerization. Acetylation is a less common form of posttranslational modification that is rarely looked at, which takes place at the var- epsilon amino group of lysines [30]. The addition of an acetyl group to lysines has a significant impact on the electrostatic properties of the protein by preventing positive charges from forming on the amino group. Acetylation can also interact with protein phosphorylation and sumoylation, as well as regulating protein stability and protecting proteins against degradation by ubiquitination. This may play a part in forming the 36-kDa TSPO dimer, which is more prominent in cancers and in response to ROS [31]. Any potential consequence of TSPO dimerization on its function and cellular distribution in breast cancer cells remains elusive. Antisera for distinct TSPO epitopes identified the main 18-kDa TSPO protein in MA-10 Leydig cells, and in the presence of hormones, immunoreactive proteins of 36 kDa were detected [31]. These antisera recognized mainly the 36-kDa protein and occasionally the 56-kDa protein in human breast cancer cells, and only limited amounts of the 18-kDa TSPO protein were observed. The higher molecular weight proteins may correspond to TSPO polymers, and their presence correlates with the higher levels of reactive oxygen species (ROS) present in breast cancer cells relative to other cells [31]. Interestingly, a rapid increase in 36-kDa TSPO dimer formation following TSA treatment was observed in MDA-MB-231 aggressive breast cancer cells, but not in MCF-7 cells (data not shown). In previous studies, we demonstrated that TSPO polymer formation is due to the formation of dityrosines as the covalent cross-linker between TSPO monomers leading to increased drug ligand binding and reduced cholesterol-binding capacity of the polymers [31]. Further addition of TSPO drug ligands to polymers increases the rate of cholesterol binding. These data indicate that ROS induce the formation of covalent TSPO polymers both in vivo and in vitro. We proposed that the TSPO polymer is the functional unit responsible for ligand-activated cholesterol binding, and that TSPO polymerization is a dynamic process modulating the function of this receptor in cholesterol transport and other cell-specific TSPO-mediated functions. In this context, human breast cancer cells appear to express only TSPO dimers, suggesting the presence of a constitutively active receptor in these cells.
The effect of TSA seems to be mediated mainly through the GC3 box, where a mutation of this site significantly reduced TSPO promoter activity in both cell lines. Sp1 and Sp3 appear to potentiate the TSA effect when co-expressed with the −121/+66 TSPO promoter in MDA-MB-231 cells, but act as inhibitors in MCF-7 cells. Sp1, Sp3, and Sp4 remained bound to the TSPO promoter following TSA treatment, indicating that the effect of TSA may not be mediated directly through binding of Sp proteins to the TSPO promoter. How TSA regulates TSPO expression remains to be investigated. TSA was not able to enhance TSPO expression in MCF-7 to levels comparable to those of MDA-MB-231, indicating that acetylation and methylation are not solely responsible for the difference in TSPO expression between the two cell lines. In addition, whether acetylation and methylation regulate TSPO expression through modifications of the intronic sequence is worthy of investigation. A database analysis of the first intron revealed the presence of multiple putative transcription factor binding sites, such as AP1, Ets, Sp1/Sp3, STAT, P300, PPARα, and c/EBP-α, among many others. The elucidation of any probable enhancing or inhibitory roles of these factors in the regulation of TSPO expression will be beneficial to understanding the mechanisms responsible for differential TSPO expression.
Analysis of the regions flanking the tss window indicated that the TSPO proximal promoter is situated within a CpG island extending approximately 470 bp upstream and 615 bp downstream. Initiation of transcription at multiple sites has been proposed to be regulated as a cassette by MED-1, a putative regulatory element identified by comparative sequence analysis of the region downstream of the transcription initiation window of several genes with TATA-less promoters [32]. No sequence motifs displaying greater than 60% identity to the MED-1 consensus sequence were found downstream of the TSPO promoter. In contrast, the flanking sequence around the common start sites at positions +24 and +38 were found to differ from the consensus mammalian initiator sequence (Inr; consensus sequence YCA+1NT/AYY) by only one and two bases, respectively. It is currently unclear whether either of these elements can reconstitute Inr function. The presence of two such elements in the same transcription window is uncommon, although TATA-less promoters with more than one Inr have been described for some genes [33, 34]. Deletion of these sequences did diminish TSPO promoter activity in MCF-7 and MDA-MB-231 cells by 20–35%, as well as in MA-10 cells (data not shown), whereas only the deletion of the +40 tss diminished promoter activity in HepG2 cells (data not shown). Interestingly, deletion of additional sequences in MCF-7 resulted in the recovery of promoter activity to maximum levels, whereas further deletion caused additional loss of activity in MDA-MB-231 cells. Substitution of unrelated sequences for these Inr-elements also affected promoter activity in MCF-7 and MDA-MB-231 cells. Both deletion and substitution mutagenesis suggested a sequence-dependence in the vicinity of the common start sites that affects promoter activity. This sequence harbors STAT1/3/5, Ets, and AhR/ARNT transcription factor binding sites, as well as a transcription start site. Interpretation of this effect is complicated, since it can disrupt the common start site at +40 and the flanking sequence of the STAT1/3/5 - Ets motif and other transcription factor binding sites. Alternatively, this mutation could disrupt an interaction that negatively regulates the TSPO promoter in MCF-7 cells. If this repressive interaction were specific to the +40 tss, it may not have been detected by deletion analysis, since the −121/+39 mutation also deletes the start site. In summary, the deregulation of factors interacting with this region as part of inducing TSPO expression is a potential mechanism by which TSPO may be up-regulated in certain cancers. Additional studies are necessary to identify the factors that interact with this region and to determine the mechanism by which these interaction(s) modulate TSPO expression.
Given its downstream location, it is possible that this region does not exert its effect at the level of transcription. Instead, this sequence could interact with trans-acting factors expressed in cells that express high levels of TSPO to alter mRNA stability or translational efficiency. If the downstream element proves to be regulated at the level of transcription, then the modular nature of the TSPO promoter should be investigated. One possible implication of the architecture of the TSPO promoter is that transcriptional regulation is directed through upstream and downstream modules that can integrate multiple signals. Activation through the array of GC boxes by Sp1 or a related protein may be sufficient to activate the TSPO promoter, but only at low to moderate levels. Based on our characterization of the TSPO promoter, it appears that interactions with GC Box 3 may activate the TSPO promoter most efficiently. However, it is likely that the other GC boxes integrate additional signals to modulate TSPO expression as part of maintaining homeostasis, much in the same way the p21 WAF/Cip1 promoter uses multiple GC boxes to integrate signals from Ras, BRCA1, and ionizing radiation [35, 36]. In this model, regulation of the TSPO promoter would require additional interactions with regulatory proteins binding to the downstream element. Full promoter activity may require interaction with an activator; however, the overlapping nature of these putative elements suggests that these downstream sequences may also contribute to cytokine-responsiveness, redox homeostasis, and tissue-specific regulation. Increased understanding of the manner in which the putative GC box module interacts with both the basal transcription machinery and the downstream regulatory module will provide important insights into the mechanism by which TSPO levels are altered in cancer and other disease states where it is overexpressed [11].
In summary, regulation of TSPO expression in MDA-MB-231 and MCF-7 cells depends on gene amplification, Sp1, Sp3 and Sp4 regulation of constitutive TSPO expression through the GC3 promoter region, and epigenetic modification of the proximal promoter, mainly at the GC3 site and the first exon, which seems to play a distinct role in mediating TSPO expression in the rich-in-TSPO MDA-MB-231 cells. Because of numerous recent reports showing that high concentrations of TSPO drug ligands inhibit proliferation of various cancer cells and sensitize cancer cells to chemotherapy [37–45], the concept of a TSPO-mediated cancer therapeutic approach has emerged. However, understanding the regulation of expression and function of TSPO in human cancer cells is paramount to any use of this protein as a drug target for cancer therapy.
Supplementary Material
Highlights.
Regulation of TSPO expression in human breast cancer cells depends on many factors.
Sp1, Sp3 and Sp4 regulation for constitutive TSPO expression through GC3 promoter region.
Epigenetic modification of the proximal promoter, at the GC3 site, and the first exon.
Differential regulation of TSPO promoter in non-aggressive and aggressive cancer cells.
Understanding its regulation provides a TSPO-mediated cancer therapeutic approach.
Acknowledgments
This work was supported by grants from the National Institutes of Health (R01 ES07747), the Susan G. Komen Breast Cancer Foundation, and a Canada Research Chair in Biochemical Pharmacology to V.P. The Research Institute of MUHC is supported by a Center grant from Le Fonds de la recherche en santé du Québec.
ABBREVIATIONS
- HBC
human breast cancer
- HMEC
human mammary epithelial cells
- TSPO
Translocator Protein (18kDa)
- ChIP
chromatin immunoprecipitation
- TSA
Trichostatin A
- AZA
5-Azacytidine
- Sp/KLF
Specificity Protein/Krüppel-like factor
Footnotes
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Dr. Amani Batarseh, Email: [email protected].
Dr. Keith D. Barlow, Email: [email protected].
Dr. Daniel B. Martinez-Arguelles, Email: [email protected].
Dr. Vassilios Papadopoulos, Email: [email protected].
References
- 1.Gavish M, Bachman I, Shoukrun R, Katz Y, Veenman L, Weisinger G, Weizman A. Enigma of the peripheral benzodiazepine receptor. Pharmacol Rev. 1999;51:629–650. [PubMed] [Google Scholar]
- 2.Papadopoulos V. Peripheral-type benzodiazepine/diazepam binding inhibitor receptor: biological role in steroidogenic cell function. Endocr Rev. 1993;14:222–240. doi: 10.1210/edrv-14-2-222. [DOI] [PubMed] [Google Scholar]
- 3.Papadopoulos V, Baraldi M, Guilarte TR, Knudsen TB, Lacapere JJ, Lindemann P, Norenberg MD, Nutt D, Weizman A, Zhang MR, Gavish M. Translocator protein (18kDa): new nomenclature for the peripheral-type benzodiazepine receptor based on its structure and molecular function. Trends Pharmacol Sci. 2006;27:402–409. doi: 10.1016/j.tips.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 4.Anholt RR, Pedersen PL, De Souza EB, Snyder SH. The peripheral-type benzodiazepine receptor. Localization to the mitochondrial outer membrane. J Biol Chem. 1986;261:576–583. [PubMed] [Google Scholar]
- 5.Culty M, Li H, Boujrad N, Amri H, Vidic B, Bernassau JM, Reversat JL, Papadopoulos V. In vitro studies on the role of the peripheral-type benzodiazepine receptor in steroidogenesis. J Steroid Biochem Mol Biol. 1999;69:123–130. doi: 10.1016/s0960-0760(99)00056-4. [DOI] [PubMed] [Google Scholar]
- 6.Garnier M, Dimchev AB, Boujrad N, Price JM, Musto NA, Papadopoulos V. In vitro reconstitution of a functional peripheral-type benzodiazepine receptor from mouse Leydig tumor cells. Mol Pharmacol. 1994;45:201–211. [PubMed] [Google Scholar]
- 7.McEnery MW, Snowman AM, Trifiletti RR, Snyder SH. Isolation of the mitochondrial benzodiazepine receptor: association with the voltage-dependent anion channel and the adenine nucleotide carrier. Proc Natl Acad Sci U S A. 1992;89:3170–3174. doi: 10.1073/pnas.89.8.3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Papadopoulos V, Boujrad N, Ikonomovic MD, Ferrara P, Vidic B. Topography of the Leydig cell mitochondrial peripheral-type benzodiazepine receptor. Mol Cell Endocrinol. 1994;104:R5–R9. doi: 10.1016/0303-7207(94)90061-2. [DOI] [PubMed] [Google Scholar]
- 9.Zoratti M, Szabo I. The mitochondrial permeability transition. Biochim Biophys Acta. 1995;1241:139–176. doi: 10.1016/0304-4157(95)00003-a. [DOI] [PubMed] [Google Scholar]
- 10.Lacapere JJ, Papadopoulos V. Peripheral-type benzodiazepine receptor: structure and function of a cholesterol-binding protein in steroid and bile acid biosynthesis. Steroids. 2003;68:569–585. doi: 10.1016/s0039-128x(03)00101-6. [DOI] [PubMed] [Google Scholar]
- 11.Batarseh A, Papadopoulos V. Regulation of translocator protein 18 kDa (TSPO) expression in health and disease states. Mol Cell Endocrinol. 2010;327:1–12. doi: 10.1016/j.mce.2010.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galiegue S, Casellas P, Kramar A, Tinel N, Simony-Lafontaine J. Immunohistochemical assessment of the peripheral benzodiazepine receptor in breast cancer and its relationship with survival. Clin Cancer Res. 2004;10:2058–2064. doi: 10.1158/1078-0432.ccr-03-0988. [DOI] [PubMed] [Google Scholar]
- 13.Han Z, Slack RS, Li W, Papadopoulos V. Expression of peripheral benzodiazepine receptor (PBR) in human tumors: relationship to breast, colorectal, and prostate tumor progression. J Recept Signal Transduct Res. 2003;23:225–238. doi: 10.1081/rrs-120025210. [DOI] [PubMed] [Google Scholar]
- 14.Hardwick M, Fertikh D, Culty M, Li H, Vidic B, Papadopoulos V. Peripheral-type benzodiazepine receptor (PBR) in human breast cancer: correlation of breast cancer cell aggressive phenotype with PBR expression, nuclear localization, and PBR-mediated cell proliferation and nuclear transport of cholesterol. Cancer Res. 1999;59:831–842. [PubMed] [Google Scholar]
- 15.Hardwick M, Cavalli LR, Barlow KD, Haddad BR, Papadopoulos V. Peripheral-type benzodiazepine receptor (PBR) gene amplification in MDA-MB-231 aggressive breast cancer cells. Cancer Genet Cytogenet. 2002;139:48–51. doi: 10.1016/s0165-4608(02)00604-0. [DOI] [PubMed] [Google Scholar]
- 16.Giatzakis C, Papadopoulos V. Differential utilization of the promoter of peripheral-type benzodiazepine receptor by steroidogenic versus nonsteroidogenic cell lines and the role of Sp1 and Sp3 in the regulation of basal activity. Endocrinology. 2004;145:1113–1123. doi: 10.1210/en.2003-1330. [DOI] [PubMed] [Google Scholar]
- 17.Quandt K, Frech K, Karas H, Wingender E, Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takai D, Jones PA. The CpG island searcher: a new WWW resource. In Silico Biol. 2003;3:235–240. [PubMed] [Google Scholar]
- 19.Carr IM, Valleley EM, Cordery SF, Markham AF, Bonthron DT. Sequence analysis and editing for bisulphite genomic sequencing projects. Nucleic Acids Res. 2007;35:e79. doi: 10.1093/nar/gkm330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu Y, Uberbacher EC. Automated gene identification in large-scale genomic sequences. J Comput Biol. 1997;4:325–338. doi: 10.1089/cmb.1997.4.325. [DOI] [PubMed] [Google Scholar]
- 21.Mertens-Talcott SU, Chintharlapalli S, Li X, Safe S. The oncogenic microRNA-27a targets genes that regulate specificity protein transcription factors and the G2-M checkpoint in MDA-MB-231 breast cancer cells. Cancer Res. 2007;67:11001–11011. doi: 10.1158/0008-5472.CAN-07-2416. [DOI] [PubMed] [Google Scholar]
- 22.Higgins KJ, Liu S, Abdelrahim M, Yoon K, Vanderlaag K, Porter W, Metz RP, Safe S. Vascular endothelial growth factor receptor-2 expression is induced by 17beta-estradiol in ZR-75 breast cancer cells by estrogen receptor alpha/Sp proteins. Endocrinology. 2006;147:3285–3295. doi: 10.1210/en.2006-0081. [DOI] [PubMed] [Google Scholar]
- 23.Wierstra I. Sp1: emerging roles--beyond constitutive activation of TATA-less housekeeping genes. Biochem Biophys Res Commun. 2008;372:1–13. doi: 10.1016/j.bbrc.2008.03.074. [DOI] [PubMed] [Google Scholar]
- 24.Kwon HS, Kim MS, Edenberg HJ, Hur MW. Sp3 and Sp4 can repress transcription by competing with Sp1 for the core cis-elements on the human ADH5/FDH minimal promoter. J Biol Chem. 1999;274:20–28. doi: 10.1074/jbc.274.1.20. [DOI] [PubMed] [Google Scholar]
- 25.Ammanamanchi S, Brattain MG. Sp3 is a transcriptional repressor of transforming growth factor-beta receptors. J Biol Chem. 2001;276:3348–3352. doi: 10.1074/jbc.M002462200. [DOI] [PubMed] [Google Scholar]
- 26.Discher DJ, Bishopric NH, Wu X, Peterson CA, Webster KA. Hypoxia regulates beta-enolase and pyruvate kinase-M promoters by modulating Sp1/Sp3 binding to a conserved GC element. J Biol Chem. 1998;273:26087–26093. doi: 10.1074/jbc.273.40.26087. [DOI] [PubMed] [Google Scholar]
- 27.Kitadai Y, Yasui W, Yokozaki H, Kuniyasu H, Haruma K, Kajiyama G, Tahara E. The level of a transcription factor Sp1 is correlated with the expression of EGF receptor in human gastric carcinomas. Biochem Biophys Res Commun. 1992;189:1342–1348. doi: 10.1016/0006-291x(92)90221-6. [DOI] [PubMed] [Google Scholar]
- 28.Braun H, Koop R, Ertmer A, Nacht S, Suske G. Transcription factor Sp3 is regulated by acetylation. Nucleic Acids Res. 2001;29:4994–5000. doi: 10.1093/nar/29.24.4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hung JJ, Wang YT, Chang WC. Sp1 deacetylation induced by phorbol ester recruits p300 to activate 12(S)-lipoxygenase gene transcription. Mol Cell Biol. 2006;26:1770–1785. doi: 10.1128/MCB.26.5.1770-1785.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Glozak MA, Sengupta N, Zhang X, Seto E. Acetylation and deacetylation of non-histone proteins. Gene. 2005;363:15–23. doi: 10.1016/j.gene.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 31.Delavoie F, Li H, Hardwick M, Robert JC, Giatzakis C, Peranzi G, Yao ZX, Maccario J, Lacapere JJ, Papadopoulos V. In vivo and in vitro peripheral-type benzodiazepine receptor polymerization: functional significance in drug ligand and cholesterol binding. Biochemistry. 2003;42:4506–4519. doi: 10.1021/bi0267487. [DOI] [PubMed] [Google Scholar]
- 32.Ince TA, Scotto KW. A conserved downstream element defines a new class of RNA polymerase II promoters. J Biol Chem. 1995;270:30249–30252. doi: 10.1074/jbc.270.51.30249. [DOI] [PubMed] [Google Scholar]
- 33.Drews VL, Lieberman AP, Meisler MH. Multiple transcripts of sodium channel SCN8A (Na(V)1.6) with alternative 5′- and 3′-untranslated regions and initial characterization of the SCN8A promoter. Genomics. 2005;85:245–257. doi: 10.1016/j.ygeno.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 34.Hoo RL, Ngan ES, Leung PC, Chow BK. Two Inr elements are important for mediating the activity of the proximal promoter of the human gonadotropin-releasing hormone receptor gene. Endocrinology. 2003;144:518–527. doi: 10.1210/en.2002-220591. [DOI] [PubMed] [Google Scholar]
- 35.Mullan PB, Quinn JE, Harkin DP. The role of BRCA1 in transcriptional regulation and cell cycle control. Oncogene. 2006;25:5854–5863. doi: 10.1038/sj.onc.1209872. [DOI] [PubMed] [Google Scholar]
- 36.Xiao H, Hasegawa T, Isobe K. Both Sp1 and Sp3 are responsible for p21waf1 promoter activity induced by histone deacetylase inhibitor in NIH3T3 cells. J Cell Biochem. 1999;73:291–302. [PubMed] [Google Scholar]
- 37.Bruce JH, Ramirez AM, Lin L, Oracion A, Agarwal RP, Norenberg MD. Peripheral-type benzodiazepines inhibit proliferation of astrocytes in culture. Brain Res. 1991;564:167–170. doi: 10.1016/0006-8993(91)91369-c. [DOI] [PubMed] [Google Scholar]
- 38.Camins A, ez-Fernandez C, Pujadas E, Camarasa J, Escubedo E. A new aspect of the antiproliferative action of peripheral-type benzodiazepine receptor ligands. Eur J Pharmacol. 1995;272:289–292. doi: 10.1016/0014-2999(94)00652-n. [DOI] [PubMed] [Google Scholar]
- 39.Garnier M, Boujrad N, Oke BO, Brown AS, Riond J, Ferrara P, Shoyab M, Suarez-Quian CA, Papadopoulos V. Diazepam binding inhibitor is a paracrine/autocrine regulator of Leydig cell proliferation and steroidogenesis: action via peripheral-type benzodiazepine receptor and independent mechanisms. Endocrinology. 1993;132:444–458. doi: 10.1210/endo.132.1.8380386. [DOI] [PubMed] [Google Scholar]
- 40.Hirsch T, Decaudin D, Susin SA, Marchetti P, Larochette N, Resche-Rigon M, Kroemer G. PK11195, a ligand of the mitochondrial benzodiazepine receptor, facilitates the induction of apoptosis and reverses Bcl-2-mediated cytoprotection. Exp Cell Res. 1998;241:426–434. doi: 10.1006/excr.1998.4084. [DOI] [PubMed] [Google Scholar]
- 41.Ikezaki K, Black KL. Stimulation of cell growth and DNA synthesis by peripheral benzodiazepine. Cancer Lett. 1990;49:115–120. doi: 10.1016/0304-3835(90)90146-o. [DOI] [PubMed] [Google Scholar]
- 42.Kupczyk-Subotkowska L, Siahaan TJ, Basile AS, Friedman HS, Higgins PE, Song D, Gallo JM. Modulation of melphalan resistance in glioma cells with a peripheral benzodiazepine receptor ligand-melphalan conjugate. J Med Chem. 1997;40:1726–1730. doi: 10.1021/jm960592p. [DOI] [PubMed] [Google Scholar]
- 43.Maaser K, Hopfner M, Jansen A, Weisinger G, Gavish M, Kozikowski AP, Weizman A, Carayon P, Riecken EO, Zeitz M, Scherubl H. Specific ligands of the peripheral benzodiazepine receptor induce apoptosis and cell cycle arrest in human colorectal cancer cells. Br J Cancer. 2001;85:1771–1780. doi: 10.1054/bjoc.2001.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neary JT, Jorgensen SL, Oracion AM, Bruce JH, Norenberg MD. Inhibition of growth factor-induced DNA synthesis in astrocytes by ligands of peripheral-type benzodiazepine receptors. Brain Res. 1995;675:27–30. doi: 10.1016/0006-8993(95)00031-k. [DOI] [PubMed] [Google Scholar]
- 45.Sanger N, Strohmeier R, Kaufmann M, Kuhl H. Cell cycle-related expression and ligand binding of peripheral benzodiazepine receptor in human breast cancer cell lines. Eur J Cancer. 2000;36:2157–2163. doi: 10.1016/s0959-8049(00)00298-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.