Abstract
Mechanical stimuli play a major role in the regulation of skeletal muscle mass, and the maintenance of muscle mass contributes significantly to disease prevention and issues associated with the quality of life. Although the link between mechanical signals and the regulation of muscle mass has been recognized for decades, the mechanisms involved in converting mechanical information into the molecular events that control this process remain poorly defined. Nevertheless, our knowledge of these mechanisms is advancing and recent studies have revealed that signaling through a protein kinase called the mammalian target of rapamycin (mTOR) plays a central role in this event. In this review we will, 1) discuss the evidence which implicates mTOR in the mechanical regulation of skeletal muscle mass, 2) provide an overview of the mechanisms through which signaling by mTOR can be regulated, and 3) summarize our current knowledge of the potential mechanisms involved in the mechanical activation of mTOR signaling.
Keywords: Contraction, Exercise, Hypertrophy, Phosphatidic Acid, Phospholipase D
Introduction
Skeletal muscles make up 40-50% of the body’s mass, and they are not only the motors that drive locomotion, but they also play a crucial role in whole body metabolism (Lee et al., 2000, Izumiya et al., 2008). With aging, skeletal muscles are a particularly important topic because a 35-40% reduction in skeletal muscle mass occurs between 20 and 80 years of age, and the loss of muscle mass is associated with disability, loss of independence and increased risk of morbidity and mortality (Proctor et al., 1998, Pahor and Kritchevsky, 1998). Accordingly, it has become well recognized that the maintenance of skeletal muscle mass contributes significantly to disease prevention and issues associated with the quality of life. Yet, despite its significance, the mechanisms involved in the regulation of muscle mass have only been vaguely defined (Seguin and Nelson, 2003).
One of the most widely recognized mechanisms for controlling muscle mass involves mechanical tension. Evidence for this mechanism comes from numerous in vivo studies which have shown that placing a chronic mechanical load on skeletal muscles results in an increase in mass, while chronic mechanical unloading results in a decrease in mass (Goldberg et al., 1975). Cell culture based studies have also demonstrated that mechanical tension can regulate muscle mass in vitro (Vandenburgh, 1987). For example, using a cell culture model, Vandenburgh et al. (1993) reported that mechanical stretch produces a large increase in protein synthesis and a smaller decrease in protein degradation, with the net result being a deposition of protein (Vandenburgh, 1987). These data suggest that, in vitro, the growth response to increased mechanical tension is due, in large part, to an increase in the rate of protein synthesis. An increased rate of protein synthesis has also been observed in numerous in vivo studies (Figure 1) and this appears to be a critical mechanism for producing growth in response to increased mechanical tension (Goldberg, 1968, Goldberg et al., 1975, Vandenburgh, 1987). Conversely, a decrease in mechanical tension (in vivo and in vitro) produces a rapid decrease in the rate of protein synthesis and this contributes to the loss of muscle mass (Vandenburgh et al., 1999, Fitts et al., 2000). Taken together, these results indicate that changes in protein synthesis play a fundamental role in the mechanical regulation of muscle mass. Hence, in order to understand how mechanical tension regulates muscle mass, it will be critical to determine how skeletal muscles sense mechanical information and convert this stimulus into the biochemical events that regulate the rate of protein synthesis.
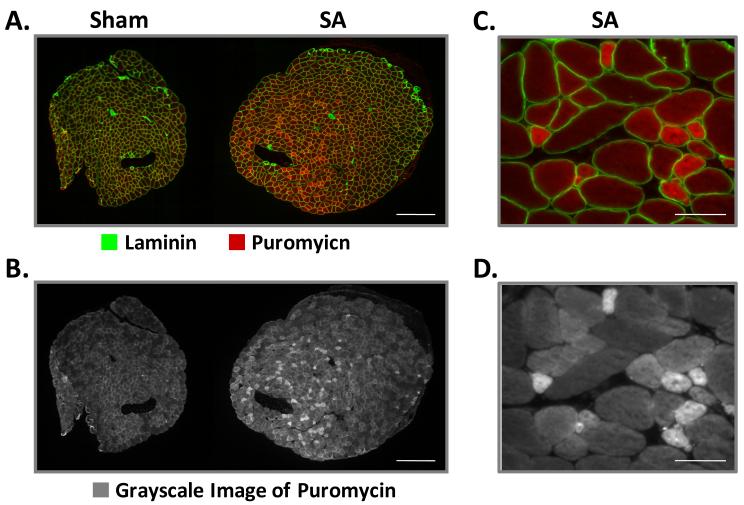
Figure 1. Chronic Mechanical Loading Induces an Increase in Protein Synthesis.
Mouse plantaris muscles were subjected to chronic mechanical loading by surgically ablating the synergist muscles (SA) as previously described (Goodman et al., 2011). For a control condition, mice were subjected to a sham surgery. At 7 days after the surgery, rates of protein synthesis were determined with the in vivo SUrface SEnsing of Translation (SUnSET) method (Goodman et al., 2011). Briefly, mice were given an injection of puromycin 30 minutes before the plantaris muscles were collected. (A-D) Cross-sections from the mid-belly of plantaris muscles subjected to sham or SA conditions were mounted adjacent to one another on a slide, and then subjected to immunohistochemistry for laminin (green) and puromycin (red). (A) Merged image of signals obtained for puromycin and laminin. (B) Grayscale image of the signal obtained for puromycin. Note: the intensity of puromycin signal reflects the rate of protein synthesis (Goodman et al., 2011). (C-D) Higher magnification image of a plantaris muscle subjected to SA reveals the presence of small fibers with very high rates of protein synthesis. The bars in A and B indicate a length of 300μm and the bars in C and D indicate a length of 40μm.
Mechanical Stimuli Induce Protein Synthesis and Hypertrophy through a Rapamycin-Sensitive Mechanism
Mechanical stimuli can regulate the rate of protein synthesis through changes in translational efficiency and/or translational capacity. Although mechanical stimuli have been shown to affect both of these processes, the primary effect of mechanical stimulation appears to occur at the level of translational efficiency (Kimball et al., 2002, Goldspink, 1977). Translational efficiency has three potential stages for regulation (initiation, elongation and termination), and mechanical stimuli appear to primarily affect the stage of initiation (Kimball et al., 2002).
Translation initiation refers to the binding of a ribosome to the mRNA, and this process is regulated by a coordinated series of biochemical events which are dependent, in part, on the 5′ structure of the mRNA. For the purpose of this review, the discussion will focus on the translation initiation of a subset of mRNAs which contain a 5′ 7-methylguanosine modification followed by an oligonucleotide tract rich in pyrimidines (5′TOP). This subset of mRNAs encodes proteins that are central to the growth process (e.g. ribosomal proteins and translation factors) (Meyuhas, 2000). Furthermore, mechanical stimuli have been shown to promote a selective increase in the proportion of 5′TOP mRNA associated with the polysomal pool, thus verifying that they are subject to translational regulation (Chen et al., 2002). For a comprehensive review on translational regulation, the reader is referred to Gebauer et al. (2004), Proud (2007) and Mahoney et al. (2009) (Proud, 2007, Gebauer and Hentze, 2004, Mahoney et al., 2009).
The translation of mRNAs that contain a 5′TOP structure are inhibited are by the drug rapamycin and one of the most intensely studied signaling proteins in the rapamycin-sensitive pathway is the ribosomal S6 kinase 1 (p70S6k). Signaling by p70S6k does not appear to be directly involved in the translation of 5′TOP mRNAs (Pende et al., 2004), but it does play a prominent role in the regulation of cell growth. For example, signaling by p70S6k is necessary for progression through the G1 checkpoint, and disruption of the p70S6k gene results in smaller muscle cells in vitro and a small mouse phenotype in vivo (Ohanna et al., 2005, Lane et al., 1993, Shima et al., 1998). It has also been shown that inhibiting signaling to p70S6k with rapamycin causes a reduction in cell size and, of particular significance to this review, rapamycin has been shown to prevent mechanically-induced changes in protein synthesis and mechanically-induced growth of skeletal muscle (Hornberger et al., 2003, Bodine et al., 2001, Fingar et al., 2002, Fingar et al., 2004, Hornberger et al., 2004, Kubica et al., 2004). Taken together, these observations suggest that components of the rapamycin-sensitive pathway, such as p70S6k, play a critical role in the mechanical regulation of protein synthesis and skeletal muscle mass.
mTOR as the Rapamycin-Sensitive Element
In vivo, rapamycin forms a complex with the immunophilin FKBP12, and this complex has been shown to bind to a protein kinase called the mammalian target of rapamycin (mTOR). The FKBP12-rapamycin complex binds to mTOR in a region termed the FRB domain. Mutating the 2035S residue in the FRB domain to 2035T inhibits the interaction of mTOR with the FKBP12-rapamycin complex and renders this mutant of mTOR (RR-mTOR) rapamycin-resistant (Lorenz and Heitman, 1995, Brown et al., 1995). For example, it has been demonstrated that over-expression of RR-mTOR in C2C12 myoblasts can rescue signaling to p70S6k from the inhibitory actions of rapamycin (Hornberger et al., 2007). Furthermore, the rescue effect is not observed when a rapamycin-resistant kinase-dead mutant of mTOR (RRKD-mTOR) is over-expressed (Hornberger et al., 2007). These observations indicate that mTOR is the rapamycin-sensitive kinase that confers signaling to p70S6k and likely numerous other downstream signaling molecules that regulate protein synthesis and skeletal muscle mass.
General Regulatory Mechanisms of mTOR Signaling
Research over the last decade has established that mTOR is the central component of a signaling network that controls cellular growth and a vast number of studies have been aimed at understanding how mTOR signaling is regulated. These studies have revealed that mTOR exists in two distinct multi-protein complexes and only one of them is inhibited by the FKBP12-rapamycin complex. The rapamycin-sensitive complex consists of the proteins mTOR, GβL and Raptor, and is collectively referred to as the mTORC1 complex (Huang and Manning, 2008, Corradetti and Guan, 2006). The other complex is referred to as the mTORC2 complex and consists of mTOR, GβL and a protein called Rictor (Guertin et al., 2006, Corradetti and Guan, 2006). Signaling by the mTORC2 complex is not directly inhibited by rapamycin, and thus, it seems unlikely that signaling by the mTORC2 complex contributes to the growth-regulatory effects that have been attributed to the rapamycin-sensitive pathway (Corradetti and Guan, 2006, Jacinto et al., 2004). Therefore, the remainder of this discussion will focus on the mechanisms that regulate signaling by the mTORC1 complex. To assist the reader, a schematic that summarizes these mechanisms is provided in Figure 2.
Figure 2. Schematic of the General Mechanisms that Regulate mTORC1 Signaling.
Signaling by mTORC1 can be activated by numerous stimuli including growth factors, nutrients and mechanical signals. In this schematic, the components of the mTORC1 complex are shaded in purple and the signaling molecules that stimulate mTORC1 signaling are shaded in green, while molecules that inhibit mTORC1 signaling are shaded in red. Dashed lines represent links in the regulatory pathways that have not been clearly established.
Phosphorylation of p70S6k on the Thr389 residue is typically used as a read-out of mTORC1 signaling, and this marker has revealed that signaling by mTORC1 can be regulated by various stimuli including growth factors, nutrients and mechanical signals. To date, the most intensely studied regulatory mechanisms of mTORC1 signaling are those which occur following stimulation with growth factors such as insulin. These studies indicate that growth factors activate class I phosphoinositide-3 kinase (PI3K) which, in-turn, promotes the activation of protein kinase B (PKB) via the 3-phosphoinositide-dependent protein kinase-1 (PDK1) (Reiling and Sabatini, 2006). Activated PKB can then phosphorylate and inhibit the activity of the tuberous sclerosis complex (TSC1/2) complex which is a GTPase-activating protein for a ras-family GTP-binding protein called Rheb (Huang and Manning, 2008). Inactivated TSC1/2 allows Rheb to become charged with GTP and, in-turn, promote the activation of mTORC1 signaling (Inoki et al., 2003). It is clear that GTP-Rheb can activate mTORC1 and it has been proposed that this might involve the sequestration of the FKBP38 protein (an endogenous inhibitor of mTORC1 signaling); however, the exact mechanism through which Rheb activates mTORC1 signaling remains a subject of intense debate (Bai et al., 2007, Ma et al., 2008, Avruch et al., 2006).
As mentioned above, nutrients such as amino acids (AA) have also been shown to play an important role in the regulation of mTORC1 signaling. Current models indicate that, unlike growth factors, nutrients activate mTORC1 through a class I PI3K-, PKB- and TSC1/2-independent mechanism (Nobukuni et al., 2005, Gulati and Thomas, 2007). However, the activation of mTORC1 by nutrients is blocked by the PI3K inhibitor wortmannin, indicating that PI3K activity is still required for this event (Nobukuni et al., 2005). This observation ultimately led to the finding that a wortmannin-sensitive class III PI3K called Vps34 might be involved in nutrient-induced mTORC1 signaling (Nobukuni et al., 2005). For example, it was reported that nutrients could induce Vps34 activity and depletion of Vps34 with siRNA blocked nutrient-induced mTORC1 signaling (Gulati and Thomas, 2007). It has also been suggested that nutrients activate Vps34 through a mechanism involving the Ca2+/calmodulin complex (CaM) (Gulati et al., 2008), however, the results of other studies have not supported this conclusion (Yan et al., 2009). The mechanism could ultimately link Vps34 to the activation of mTORC1 is even less clear, but might involve enhanced binding of Rheb to mTORC1; however, it has also been reported that nutrient stimulation does not alter the amount of GTP bound Rheb (Nobukuni et al., 2005, Long et al., 2005). Furthermore, genetic studies in Drosophilia and C. elegans have recently challenged the potential role of Vps34 in the regulation of TORC1 signaling (Juhasz et al., 2008, Avruch et al., 2009). Thus, there is still much to be disputed with regards to the potential role of Vps34 in amino acid induced mTORC1 signaling.
Currently, the most widely accepted mechanism for explaining how amino acids regulate mTORC1 signaling involves the Rag subfamily of Ras small GTPases. Mammalian cells express four Rag GTPases (RagA, RagB, RagC and RagD) and two independent groups have demonstrated that Rag GTPases are necessary for the activation of mTORC1 signaling by amino acids (Kim et al., 2008, Sancak et al., 2008). Furthermore, both groups demonstrated that overexpression of constitutively GTP bound RagA or RagB is sufficient to induce an increase in mTORC1 signaling and an increase in the size of both Drosophilia and mammalian cells (Kim et al., 2008, Sancak et al., 2008). More recently, amino acids have been shown to induce translocation of mTORC1 to lysosomal membranes where both the Rag GTPases and Rheb reside (Sancak et al.). Furthermore, the lysosomal membrane recruitment of mTORC1 has been shown to be dependent on a complex of proteins encoded by the MAPKSP1, ROBLD3 and c11orf59 genes, and this complex has been termed the Ragulator complex (Sancak et al.). Of particular significance, constitutive targeting of mTORC1 to the lysosomal surface renders mTORC1 signaling independent of amino acids, Rag GTPases and the Ragulator complex, but it remains dependent on Rheb (Sancak et al.). Thus, the interaction of mTORC1 with Rheb, via a Rag GTPase-Ragulator dependent translocation of mTORC1 to lysosomal membranes, appears to be a critical event for the activation of mTORC1 signaling in response to amino acids and possibly several other stimuli.
Another important regulator of mTORC1 signaling is the AMP-activated protein kinase (AMPK). During periods of energy deprivation (i.e. exercise, hypoxia and nutrient deprivation), AMP levels rise and promote the activation of AMPK (Kimball, 2006, Musi et al., 2003, Fujii et al., 2006, Kim et al., 2004). Activated AMPK can phosphorylate and activate the TSC1/2 complex which represses the Rheb-induced activation of mTORC1 signaling. For this reason, it has been proposed that excess nutrients may indirectly activate mTORC1 signaling through repression of the inhibitory signals provided by the AMPK → TSC1/2 -| Rheb → mTORC1 pathway (Hahn-Windgassen et al., 2005). Furthermore, it was recently demonstrated that AMPK can directly phosphorylate raptor, and that this event inhibits mTORC1 signaling (Gwinn et al., 2008). Thus, AMPK appears to play an important role in the regulation of mTORC1 signaling and the exact mechanisms involved in this process continue to be defined.
An additional regulatory mechanism of mTORC1 signaling involves the lipid second messenger phosphatidic acid (PA) (Foster, 2007). The potential role of PA in the regulation of mTORC1 signaling was revealed in a study which demonstrated that PA can directly bind to mTOR in a region that lies within the FRB domain and, in doing so, activates mTORC1 signaling (Fang et al., 2001). Furthermore, there is evidence that PA competes with the FKBP12-rapamycin complex for binding to the FRB domain (Fang et al., 2001, Chen et al., 2003, Veverka et al., 2008). Based on these observations, it has been proposed that rapamycin inhibits mTORC1 signaling by removing PA from the FRB domain. Since these initial reports, several additional studies have provided evidence that PA plays a fundamental role in the regulation of mTORC1 signaling. For example, over-expression of diacylglycerol kinase, an enzyme that converts diacylglycerol into PA, induces the activation of mTORC1 signaling and this event is blocked when the PA binding domain of mTOR is mutated (Avila-Flores et al., 2005). Over-expression of lysophosphatidic acid acyltransferase, an enzyme that converts lysophosphatidic acid into PA, has also been shown to activate mTORC1 signaling (Tang et al., 2006). Finally, it was recently suggested that PA might be the primary effector of Rheb that promotes the activation of mTORC1 signaling (Sun et al., 2008). This hypothesis was based on the findings that GTP-bound Rheb binds to, and activates, phospholipase D (PLD), an enzyme that generates PA from phosphotidylcholine (Sun et al., 2008). Furthermore, it was reported that knocking down the expression of PLD prevented the activation of mTORC1 by Rheb (Sun et al., 2008). Thus, several lines of evidence suggest that PA is a direct regulator of mTORC1 signaling, and therefore, understanding the exact role that PA plays in the regulation of mTORC1 will be an important area for future studies.
The work cited above demonstrates that the regulation of mTORC1 signaling is controlled by a complex network of signaling events, and our understanding of this network is rapidly evolving. Due to page limitations, several other potentially important regulatory mechanisms have not been discussed and hence, the reader is referred to Yang et al. (2007), Foster (2007), Kim and Guan (2009) and Avruch et al. (2009) for more comprehensive reviews on the mechanisms that regulate mTORC1 signaling (Yang and Guan, 2007, Foster, 2007, Kim and Guan, 2009, Avruch et al., 2009).
The Role of mTORC1 in the Regulation of Skeletal Muscle Mass by Mechanical Stimuli
In 1999, a landmark study by Baar and Esser demonstrated that resistance-exercise promotes an increase in p70S6k phosphorylation, and the magnitude of the increase in p70S6k phosphorylation was highly correlated with the concomitant hypertrophic response of the muscle (Baar and Esser, 1999). In 2001, Bodine et al. extended these findings by using a model of chronic mechanical loading to demonstrate that rapamycin prevents the mechanically-induced increase p70S6k phosphorylation. More importantly, this study also demonstrated that rapamycin blocked the hypertrophic response in mechanically loaded muscles (Bodine et al., 2001). Others have since repeated these observations and also demonstrated that rapamycin prevents the increase in protein synthesis that occurs in response to mechanical stimulation (Hornberger et al., 2004, Hornberger et al., 2003, Drummond et al., 2009, Kubica et al., 2005). Taken together, these studies suggest that mechanical stimulation is sufficient for the activation of mTORC1 signaling, and that signaling through a rapamycin-sensitive mechanism is necessary for mechanically-induced changes in protein synthesis and compensatory growth.
The Role of PI3K/PKB in the Regulation of mTORC1 Signaling and Muscle Mass by Mechanical Stimuli
To date, signaling through the rapamycin-sensitive pathway (presumably involving mTORC1) is the only molecular pathway that has repeatedly been identified as being necessary for mechanically-induced growth of skeletal muscle. Since rapamycin is considered to be a highly specific inhibitor of mTORC1, it has been widely concluded that signaling through mTORC1 is necessary for mechanically-induced growth. However, genetic evidence to support this fundamental conclusion is lacking. Despite this limitation, numerous investigations have been aimed at defining how mechanical stimuli activate mTORC1 signaling. The most widely discussed model that has emerged from these studies is that mechanical stimuli activate mTORC1 through a mechanism involving the insulin-like growth factor 1 (IGF-1) and the PI3K→PKB→mTORC1 pathway. Support for this model has come from three primary observations; i) mechanical stimulation promotes an increase in IGF-1 content (Note: IGF-1 has been shown to activate the PI3K→PKB→mTORC1 pathway and induce skeletal muscle growth) (Adams and Haddad, 1996, Adams and McCue, 1998, Rommel et al., 2001), ii) mechanical stimulation activates signaling through PI3K and PKB (Bolster et al., 2003, Hornberger et al., 2003, Sakamoto et al., 2003, Atherton et al., 2005, Deshmukh et al., 2006, Haddad and Adams, 2006, Bodine et al., 2001, Carlson et al., 2001, Spangenburg and McBride, 2006, Lockhart et al., 2006, Bruss et al., 2005), and iii) constitutive activation of the PI3K→PKB pathway induces mTORC1 signaling and promotes skeletal muscle growth via a rapamycin-sensitive mechanism (Varma and Khandelwal, 2007, Bodine et al., 2001, Pallafacchina et al., 2002). Combined, these observations provide a solid rationale for the hypothesis that mechanical stimuli activate mTORC1 signaling and growth through a mechanism involving IGF-1 and the PI3K→PKB→mTORC1 pathway; however, recent studies have challenged the validity of this hypothesis.
The first study that challenges the above hypothesis comes from Spangenburg et al. 2008 (Spangenburg et al., 2008). In this study, a transgenic mouse that expresses a dominant negative IGF-1 receptor specifically in skeletal muscle was employed. It was demonstrated that skeletal muscles from these mice fail to show a signaling response (increase in PKB phosphorylation) following IGF-1 stimulation; however, the hypertrophic response induced by mechanical-overload was fully preserved in muscles from these mice. These observations led to the conclusion that a functional IGF-1 receptor is not required for mechanically-induced growth of skeletal muscle.
Another piece of evidence that challenges the IGF-1 / PI3K→PKB→mTORC1 model comes from a study that evaluated the temporal response of signaling through PI3K→PKB and mTORC1 following a bout of eccentric contractions (O’Neil et al., 2009). In this study, phosphorylation of PKB on the serine 473 residue [P-PKB] was used as a marker of signaling through the PI3K→PKB pathway, and phosphorylation of p70S6k on the threonine 389 residue [P-p70(389)] was used as a marker of signaling through mTORC1. Consistent with other studies (Spangenburg and McBride, 2006, Lockhart et al., 2006), eccentric contractions induced signaling through PI3K→PKB; however; this was a very transient event (<15 minutes) and markedly different from the activation of mTORC1 signaling which was sustained for a long duration (>12 hours) (O’Neil et al., 2009). Thus, there does not appear to be a temporal relationship between the activation of PI3K→PKB and mTORC1 following eccentric contractions. Furthermore, a number of studies have shown that pharmacological inhibition of PI3K with wortmannin, or genetic deletion of PKB1, does not block the mechanical activation of mTORC1 signaling (Hornberger et al., 2007, O’Neil et al., 2009, Hornberger et al., 2004). In other words, there are several lines of evidence which indicate that mechanical stimuli activate mTORC1 signaling through a PI3K/PKB-independent mechanism.
The observation that mechanical stimuli induce mTORC1 signaling through a PI3K/PKB-independent mechanism raises the question of whether a PI3K/PKB-independent activation of mTORC1 is actually sufficient to induce skeletal muscle growth. The importance of this question is highlighted by previous studies which have shown that constitutive activation of PI3K/PKB induces growth, and that this occurs through a rapamycin-sensitive mechanism. However, activation of PI3K/PKB can also induce signaling through a variety of mTORC1-independent growth regulatory molecules including GSK3β and the FOXO transcription factors (Nader, 2005, Sandri, 2008, Frost and Lang, 2007). Thus, based on these studies, it was not clear if mTORC1 signaling is sufficient, or simply permissive, for PI3K/PKB-induced growth. In an effort to address this issue, we recently used transient over-expression of Rheb as a means to induce a PI3K/PKB-independent activation of mTORC1 in adult mouse skeletal muscles. The results of this study demonstrated that over-expression of Rheb was indeed sufficient for the induction of a rapamycin-sensitive hypertrophic response (Goodman et al., 2010). Furthermore, transgenic mice with muscle specific expression of various mTOR mutants were used to demonstrate that mTOR is the rapamycin-sensitive element that confers the hypertrophic effects of Rheb and that the kinase activity of mTOR is necessary for this event (Goodman et al., 2010). Combined, these results provided direct genetic evidence that a PI3K/PKB-independent activation of mTORC1 signaling is sufficient to induce hypertrophy. Thus, it can now be argued that IGF-1 and the activation of the PI3K/PKB pathway are sufficient for the induction of skeletal muscle growth, but these mechanisms do not appear to be necessary for the activation of mTORC1 signaling or the growth that occurs in response to mechanical stimuli.
The Potential Role of Stretch Activated Ion Channels in the Mechanical Activation of mTORC1 Signaling
Based on the studies cited above, it appears that mechanical stimuli activate mTORC1 signaling through a PI3K/PKB-independent mechanism, but the exact identity of the PI3K/PKB-independent mechanism has not been defined. In one study aimed at identifying this mechanism, gadolinium and streptomycin were used to inhibit stretch activated ion channels (SACs). The results demonstrated that both gadolinium and streptomycin can partially block the eccentric contraction induced increase in mTORC1 signaling (Spangenburg and McBride, 2006). These observations are intriguing, but it is not clear how the flux of ions through SACs would regulate mTORC1 signaling. One possibility is that an influx of calcium through SACs would activate the CaM complex / Vps34 pathway along with concomitant activation of mTORC1. However, it has also been shown that the eccentric contractions can induce mTORC1 signaling in the absence of changes in Vps34 activity (MacKenzie et al., 2009). Furthermore, it has been demonstrated that an intracellular chelator of calcium (BAPTA-AM) does not block the mechanical activation of mTORC1 signaling (Hornberger et al., 2006). Thus, further studies are needed to define the potential link between SACs and the mechanical activation of mTORC1 signaling.
The Potential Role of Amino Acids in the Mechanical Activation of mTORC1 Signaling
As mentioned above, amino acids have been implicated in the regulation of mTORC1 signaling, and unlike growth factors, amino acids do not activate signaling through PI3K/PKB (Kimball et al., 1999, Armstrong et al., 2001). Furthermore, mechanical stimuli have been shown to induce an increase in amino acid uptake (Vandenburgh and Kaufman, 1981, MacKenzie et al., 2009). Based on these points it has been proposed that amino acids might be part of the PI3K/PKB-independent mechanism through which mechanical stimuli activate mTORC1 signaling. However, it has also been reported that the activation of mTORC1 following eccentric contractions precedes the increase in intracellular amino acids (MacKenzie et al., 2009). Furthermore, in isolated skeletal muscles, mechanical stimuli can activate mTOR in the absence of exogenous amino acids (Hornberger and Chien, 2006). Finally, it has also been shown that inhibition of PI3K with wortmannin prevents the activation of mTORC1 signaling by amino acids but not mechanical stimuli (Hornberger and Chien, 2006). Thus, mechanical stimuli appear to activate mTORC1 signaling via a mechanism that is distinct from the PI3K-dependent mechanism employed by amino acids.
The Potential Role of PLD in the Mechanical Activation of mTORC1 Signaling
Previous studies have demonstrated that a phospholipase C (PLC) / phospholipase D (PLD) inhibitor (neomycin) can prevent the mechanical activation of mTORC1 signaling (Hornberger et al., 2006). Furthermore, it has been demonstrated that inhibition of PLC with U73122 does not inhibit the mechanical activation of mTORC1 signaling (Hornberger et al., 2006). Combined, these observations suggest that the effects of neomycin are most likely attributable to the inhibition of PLD. In support of this conclusion, studies employing 1-butanol, a primary alcohol that inhibits PLD-catalyzed PA formation, and 2-butanol, a secondary alcohol that does not inhibit PLD-catalyzed PA formation, have demonstrated that 1-butanol can block the activation of mTORC1 signaling by passive stretch and eccentric contractions, but 2-butanol has no effect (Hornberger et al., 2006, Fang et al., 2001, Facchinetti et al., 1998, Wakelam et al., 1995, Shen et al., 2001, Hornberger et al., 2007, O’Neil et al., 2009). Based on these results, it would appear that the synthesis of PA by PLD is necessary for the mechanical activation of mTORC1 signaling. However, recent studies have shown that some of 1-butanol’s biological effects cannot be explained by the inhibition of PLD activity (Su et al., 2009). Thus, further studies will be needed to clarify the potential role of PLD in the mechanical activation of mTORC1 signaling.
Mechanical Stimulation Promotes an Increase in PA
Based on the results described above, it was proposed that mechanical stimuli induce an increase in PA concentration ([PA]), and that this occurs through a PLD-dependent mechanism. Consistent with this hypothesis, several studies have shown that mechanical stimuli can induce an increase in [PA] (O’Neil et al., 2009, Hornberger et al., 2006, Cleland et al., 1989). It has also been shown that elevations in [PA] occur specifically in response to contractions that induce growth and mTORC1 signaling (O’Neil et al., 2009). In addition, both neomycin and 1-butanol can prevent the mechanically-induced increase in [PA] (Hornberger et al., 2006, O’Neil et al., 2009). Combined, these results indicate that mechanical stimulation induces an increase in [PA], and the data with neomycin and 1-butanol suggest that PLD activity may be necessary for this event.
Enhanced Binding of PA to mTOR in Response to Mechanical Stimulation
As mentioned previously, PA has been reported to directly activate mTORC1 signaling by binding to mTOR on its FRBs domain (Fang et al., 2001, Veverka et al., 2008). The FRB domain is also the site where the FKBP12-rapamycin complex binds to mTOR and inhibits mTORC1 signaling (Fang et al., 2001, Veverka et al., 2008). The overlap of these binding sites suggests that the FKBP12-rapamycin complex and PA compete with one another for binding to the FRB domain (Fang et al., 2001, Chen et al., 2003, Veverka et al., 2008). Therefore, enhanced binding of PA to mTOR would be expected to confer resistance to the inhibitory effects that rapamycin has on mTORC1 signaling. This is an important point because previous studies have demonstrated that the half maximal inhibitory concentration (IC50) of rapamycin for inhibiting p70S6k(389) phosphorylation in mechanically stimulated muscles is significantly higher than the IC50 that is found in control muscles (Hornberger et al., 2006). The higher IC50 in mechanically stimulated muscles indicates that mechanical stimulation confers resistance to rapamycin; an observation that is consistent with a model in which the mechanical activation of mTORC1 signaling results from enhanced binding of PA to the FRB domain in mTOR.
PA can Induce mTORC1 Signaling via a PI3K-Independent Mechanism
The results presented so far provide reasonable evidence in favor of the hypothesis that an increase in [PA] is responsible for the mechanical activation of mTORC1 signaling. In further support of this hypothesis, it has been shown that incubating C2C12 myoblasts with exogenous PA is sufficient to induce a rapid (< 20min) increase in mTORC1 signaling (O’Neil et al., 2009). In the same study, it was also shown that, similar to mechanical stimuli, exogenous PA induced mTORC1 signaling through a PI3K-independent mechanism. These results are important, however, they should be interpreted with caution because exogenous PA can easily be metabolized and converted to other lipids. For example, unlike what has been shown in C2C12 myoblasts, a recent study is Rat2 fibroblasts reported that exogenous PA does not induce a rapid activation of mTORC1 signaling (Winter et al.). Instead, in this study, activation of mTORC1 signaling was not observed until 2hr after the addition of the exogenous PA, and it was suggested that the delayed activation of mTORC1 signaling resulted from the conversion of the exogenous PA to lysophosphatidic acid (Winter et al.). The reasons why exogenous PA did not induce a rapid activation of mTORC1 signaling in this study are less clear, but may have been due to the species of PA employed or the experimental procedures that were used to preparing the PA.
Changes in PLD Activity are not Sufficient for a Mechanically-Induced Increase in [PA]
There is a growing body of evidence which implicates PA in the mechanical regulation of mTORC1 signaling; however, the results from studies aimed at identifying the enzymes that promote the increase in PA are less clear. For example, it has been demonstrated that both neomycin and 1-butanol can prevent the mechanically-induced increase in [PA]. If it is assumed that the effects of neomycin and 1-butanol are due to the inhibition of PLD activity, then these results would indicate that PLD activity is necessary for the mechanically-induced increase in [PA]. But, as mentioned above, concerns about the non-specific effects of 1-butanol have recently been raised, and the same concerns are probably equally valid for neomycin. Furthermore, an increase in PLD activity does not appear to be sufficient for the increase in PA. For example, it has been demonstrated that PLD activity is elevated during the first 15min following the onset of mechanical stimulation, after which point it returns back to, or slightly below, control levels (Hornberger et al., 2006). On the other hand, mechanical stimulation promotes a progressive increase in [PA] throughout the first 30min after the onset of mechanical stimulation, after which point, [PA] reaches an elevated steady-state (Hornberger et al., 2006). Specifically, in mechanically stimulated muscles, [PA] is 138% greater than control muscles at 15min after the initiation of mechanical stimulation and this value increases to 187% of control at 30min after the initiation of mechanical stimulation. Yet, when compared with time matched control muscles, PLD activity during the 15-30min time period is 27% lower in mechanically stimulated muscles (P = 0.06) (Hornberger et al., 2006). Based on these observations, it can be argued that PLD might contribute to the very early part of the mechanically-induced increase in [PA], but it is not sufficient for the majority of the increase in [PA] that occurs in response to mechanical stimulation. Thus, the enzymes responsible for promoting the mechanically-induced increase in [PA] remain to be defined.
Potential Regulators of PA / mTORC1 Signaling
As highlighted above, PA has emerged as a potentially direct regulator of mTORC1 signaling. To date, five distinct classes of enzymes have been implicated in the regulation of [PA]. For example, PA can be synthesized from phosphotidylcholine (PC) by phospholipase D (PLD), from lysophosphatidic acid (LPA) by lysophosphatidic acid acyltransferases (LPAAT), and from diacylglycerol (DAG) by diacylglycerol kinases (DAGK) (Foster, 2007, Wang et al., 2006). Furthermore, [PA] can be controlled by enzymes that degrade PA which includes the conversion of PA to LPA by A type phospholipases (PLA), and the conversion of PA to DAG by phosphatidic acid phosphatases (PAP) (Wang et al., 2006, Aoki et al., 2007, Carman and Han, 2006). A schematic summarizing these enzymatic pathways and a review of the individual components is provided below (Figure 3).
Figure 3.
Schematic Overview of the Lipids and Enzymatic Pathways that Regulate [PA] and Potentially mTORC1 Signaling.
The most highly studied enzyme that regulates [PA] is PLD, and two PLD genes (PLD1 and PLD2) have been identified in skeletal muscle (Kodaki and Yamashita, 1997, Katayama et al., 1998). To date, several studies have shown that over-expression of PLD2 induces mTORC1 signaling and the lipase activity of PLD2 is required for this event (Chen et al., 2005, Ha et al., 2006). Thus, under the appropriate circumstance, the synthesis of PA by PLD can be an important regulatory mechanism of mTORC1 signaling. For a comprehensive review on PLD see Jenkins et al. (2005) and Oude Weernink et al. (2007) (Jenkins and Frohman, 2005, Oude Weernink et al., 2007)
Another well studied regulator of [PA] is DAGK. DAGK phosphorylates DAG to form PA and at least 10 isoforms of this enzyme have been identified (Sakane et al., 2007). Of the DAGK isoforms, DAGKα, δ, ε and ζ appear to be most highly expressed in skeletal muscle (Takahashi et al., 2007). As mentioned above, over-expression of DAGKζ induces mTORC1 signaling and this event is blocked when the PA binding domain of mTOR is mutated (Avila-Flores et al., 2005). Thus, the generation of PA by DAGK also appears to be an important mechanism in the regulation of mTORC1 signaling. For a further review on DAGKs see Topham (2006) and Sakane et al. (2007) (Topham, 2006, Sakane et al., 2007).
The LPAATs, also known as the 1-acyl-sn-glycerol 3-phosphate acyltransferases (AGPAT), are another potentially important regulator of [PA] and mTORC1 signaling. These enzymes catalyze the acylation of LPA to form PA and at least nine isoforms of LPAAT have been identified, all of which appear to be expressed in skeletal muscle (Lu et al., 2005). It is worth noting that pharmacological inhibition of LPAAT activity has been shown to block the activation of mTORC1 signaling by Angiotensin II, and it prevents myoblast proliferation (Coon et al., 2003). Furthermore, as noted above, over-expression of LPAAT has been shown to induce mTORC1 signaling (Tang et al., 2006). These observations are all consistent with a potentially important role for LPAAT in the regulation of [PA] and mTORC1 signaling.
The [PA] is not only regulated by enzymes that synthesize PA, but also by enzymes that degrade PA. The most highly studied class of enzymes that degrade PA is the PLAs. The PLAs are a very diverse class of enzymes which can be further divided into two major subgroups (PLA1 and PLA2). PLA1 catalyzes the hydrolysis of the sn-1 position of glycerophospholipids such as PA and nine isoforms of this enzyme have been identified (Aoki et al., 2007). On the other hand, PLA2 catalyzes the hydrolysis of the sn-2 position of glycerophospholipids and nineteen isoforms of PLA2 have been identified (Murakami and Kudo, 2002). Currently, we are not aware of any studies which have addressed the potential role of PLAs in the regulation of mTORC1 signaling; however, it is worth noting that deletion of PLA2g4a, a gene that encodes a PLA2 isoform, was recently shown to induce skeletal muscle hypertrophy and augment pressure overload induced growth of the heart. Although this study did not address the potential role of PA and mTORC1 in these phenotypes, it is tempting to speculate that the enhanced growth may have resulted from an accumulation of PA that would have normally been degraded by this enzyme (Haq et al., 2003).
The other important class of enzymes that degrade PA are the phosphatidic acid phosphatases (PAP) and these enzymes can be separated into two distinct subgroups (PAP1 and PAP2) (Carman and Han, 2006). PAP1s convert newly synthesized PA to DAG and a protein called Lipin 1 accounts for the majority of PAP1 activity in skeletal muscle (Harris et al., 2007, Nanjundan and Possmayer, 2003). Unlike PAP1, PAP2s can degrade a variety of lipid phosphates including PA, sphingosine-1-phosphate and ceramide-1-phosphate (Carman and Han, 2006). For this reason, PAP2 has generally been referred to as lipid phosphate phosphatase (Nanjundan and Possmayer, 2003). At least three isoforms of PAP2 have been identified, and all of them appear to be expressed in skeletal muscle (Kai et al., 1997, Hooks et al., 1998, Carman and Han, 2006). Previous studies have also demonstrated that pharmacological inhibition of the PAPs can induce an increase in [PA] and mTORC1 signaling in skeletal muscle (Hornberger et al., 2006). Thus, PAPs are likely to play an important role in the regulation of mTORC1 signaling and will likely be an important area of future studies.
Conclusions
It is clear that mechanical stimuli play a major role in the regulation of muscle mass. Although the link between mechanical signals and the regulation of muscle mass has been recognized for decades, the mechanisms involved in converting mechanical signals into the molecular events that control this process have only been vaguely defined. Nevertheless, our knowledge of these mechanisms is advancing and several lines of evidence have led to the conclusion that signaling by mTORC1 plays a central role in this event. Furthermore, it appears that mechanical stimuli activate mTORC1 signaling through a PI3K-independent mechanism and there is growing body of evidence which implicates PA in this process. However, additional studies are needed to fully establish the role of PA in the mechanical regulation of mTORC1 signaling and skeletal muscle mass. Furthermore, the enzymes responsible for the mechanically-induced increase in PA remain to be determined and will be an important area of focus for future studies.
Acknowledgements
This work was supported by NIH grant AR057347
Abbreviations
- AGPAT
1-acyl-sn-glycerol 3-phosphate acyltransferase
- AMPK
AMP-activated protein kinase
- CaM
Ca2+/calmodulin complex
- DAG
diacylglycerol
- DAGK
diacylglycerol kinase
- IC50
maximal inhibitory concentration
- IGF-1
insulin-like growth factor 1
- LPA
lysophosphatidic acid
- LPAAT
lysophosphatidic acid acyltransferase
- mTOR
mammalian target of rapamycin
- mTORC1
mTOR complex 1
- mTORC2
mTOR complex 2
- PA
phosphatidic acid
- PAP
phosphatidic acid phosphatases
- PC
phosphotidylcholine
- PDK1
3-phosphoinositide-dependent protein kinase-1
- PI3K
phosphoinositide-3 kinase
- PKB
protein kinase B
- PLA
A type phospholipase
- PLC
phospholipase C
- PLD
phospholipase D
- p70S6k
ribosomal S6 kinase 1
- Rheb
ras homologue enriched in brain
- RR-mTOR
rapamycin-resistant mutant of mTOR
- RRKD-mTOR
rapamycin-resistant kinase dead mutant of mTOR
- SACs
stretch activated ion channels
- TSC1/2
tuberous sclerosis complex
- 5′TOP
5′ 7-methylguanosine modification followed by an oligonucleotide tract rich in pyrimidines
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams GR, Haddad F. The relationships among IGF-1, DNA content, and protein accumulation during skeletal muscle hypertrophy. J Appl Physiol. 1996;81:2509–2516. doi: 10.1152/jappl.1996.81.6.2509. [DOI] [PubMed] [Google Scholar]
- Adams GR, McCue SA. Localized infusion of IGF-I results in skeletal muscle hypertrophy in rats. J Appl Physiol. 1998;84:1716–1722. doi: 10.1152/jappl.1998.84.5.1716. [DOI] [PubMed] [Google Scholar]
- Aoki J, Inoue A, Makide K, Saiki N, Arai H. Structure and function of extracellular phospholipase A1 belonging to the pancreatic lipase gene family. Biochimie. 2007;89:197–204. doi: 10.1016/j.biochi.2006.09.021. [DOI] [PubMed] [Google Scholar]
- Armstrong JL, Bonavaud SM, Toole BJ, Yeaman SJ. Regulation of glycogen synthesis by amino acids in cultured human muscle cells. J Biol Chem. 2001;276:952–956. doi: 10.1074/jbc.M004812200. [DOI] [PubMed] [Google Scholar]
- Atherton PJ, Babraj J, Smith K, Singh J, Rennie MJ, Wackerhage H. Selective activation of AMPK-PGC-1alpha or PKB-TSC2-mTOR signaling can explain specific adaptive responses to endurance or resistance training-like electrical muscle stimulation. FASEB J. 2005;19:786–788. doi: 10.1096/fj.04-2179fje. [DOI] [PubMed] [Google Scholar]
- Avila-Flores A, Santos T, Rincon E, Merida I. Modulation of the mammalian target of rapamycin pathway by diacylglycerol kinase-produced phosphatidic acid. J Biol Chem. 2005;280:10091–10099. doi: 10.1074/jbc.M412296200. [DOI] [PubMed] [Google Scholar]
- Avruch J, Hara K, Lin Y, Liu M, Long X, Ortiz-Vega S, Yonezawa K. Insulin and amino-acid regulation of mTOR signaling and kinase activity through the Rheb GTPase. Oncogene. 2006;25:6361–6372. doi: 10.1038/sj.onc.1209882. [DOI] [PubMed] [Google Scholar]
- Avruch J, Long X, Ortiz-Vega S, Rapley J, Papageorgiou A, Dai N. Amino acid regulation of TOR complex 1. Am J Physiol Endocrinol Metab. 2009;296:E592–602. doi: 10.1152/ajpendo.90645.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baar K, Esser K. Phosphorylation of p70(S6k) correlates with increased skeletal muscle mass following resistance exercise. Am J Physiol. 1999;276:C120–127. doi: 10.1152/ajpcell.1999.276.1.C120. [DOI] [PubMed] [Google Scholar]
- Bai X, Ma D, Liu A, Shen X, Wang QJ, Liu Y, Jiang Y. Rheb activates mTOR by antagonizing its endogenous inhibitor, FKBP38. Science. 2007;318:977–980. doi: 10.1126/science.1147379. [DOI] [PubMed] [Google Scholar]
- Bodine SC, Stitt TN, Gonzalez M, Kline WO, Stover GL, Bauerlein R, Zlotchenko E, Scrimgeour A, Lawrence JC, Glass DJ, Yancopoulos GD. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat Cell Biol. 2001;3:1014–1019. doi: 10.1038/ncb1101-1014. [DOI] [PubMed] [Google Scholar]
- Bolster DR, Kubica N, Crozier SJ, Williamson DL, Farrell PA, Kimball SR, Jefferson LS. Immediate response of mammalian target of rapamycin (mTOR)-mediated signalling following acute resistance exercise in rat skeletal muscle. J Physiol. 2003;553:213–220. doi: 10.1113/jphysiol.2003.047019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown EJ, Beal PA, Keith CT, Chen J, Shin TB, Schreiber SL. Control of p70 s6 kinase by kinase activity of FRAP in vivo. Nature. 1995;377:441–446. doi: 10.1038/377441a0. [DOI] [PubMed] [Google Scholar]
- Bruss MD, Arias EB, Lienhard GE, Cartee GD. Increased phosphorylation of Akt substrate of 160 kDa (AS160) in rat skeletal muscle in response to insulin or contractile activity. Diabetes. 2005;54:41–50. doi: 10.2337/diabetes.54.1.41. [DOI] [PubMed] [Google Scholar]
- Carlson CJ, Fan Z, Gordon SE, Booth FW. Time course of the MAPK and PI3-kinase response within 24 h of skeletal muscle overload. J Appl Physiol. 2001;91:2079–2087. doi: 10.1152/jappl.2001.91.5.2079. [DOI] [PubMed] [Google Scholar]
- Carman GM, Han GS. Roles of phosphatidate phosphatase enzymes in lipid metabolism. Trends Biochem Sci. 2006;31:694–699. doi: 10.1016/j.tibs.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Rodrik V, Foster DA. Alternative phospholipase D/mTOR survival signal in human breast cancer cells. Oncogene. 2005;24:672–679. doi: 10.1038/sj.onc.1208099. [DOI] [PubMed] [Google Scholar]
- Chen Y, Zheng Y, Foster DA. Phospholipase D confers rapamycin resistance in human breast cancer cells. Oncogene. 2003;22:3937–3942. doi: 10.1038/sj.onc.1206565. [DOI] [PubMed] [Google Scholar]
- Chen YW, Nader GA, Baar KR, Fedele MJ, Hoffman EP, Esser KA. Response of rat muscle to acute resistance exercise defined by transcriptional and translational profiling. J Physiol. 2002;545:27–41. doi: 10.1113/jphysiol.2002.021220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleland PJ, Appleby GJ, Rattigan S, Clark MG. Exercise-induced translocation of protein kinase C and production of diacylglycerol and phosphatidic acid in rat skeletal muscle in vivo. Relationship to changes in glucose transport. J Biol Chem. 1989;264:17704–17711. [PubMed] [Google Scholar]
- Coon M, Ball A, Pound J, Ap S, Hollenback D, White T, Tulinsky J, Bonham L, Morrison DK, Finney R, Singer JW. Inhibition of lysophosphatidic acid acyltransferase beta disrupts proliferative and survival signals in normal cells and induces apoptosis of tumor cells. Mol Cancer Ther. 2003;2:1067–1078. [PubMed] [Google Scholar]
- Corradetti MN, Guan KL. Upstream of the mammalian target of rapamycin: do all roads pass through mTOR? Oncogene. 2006;25:6347–6360. doi: 10.1038/sj.onc.1209885. [DOI] [PubMed] [Google Scholar]
- Deshmukh A, Coffey VG, Zhong Z, Chibalin AV, Hawley JA, Zierath JR. Exercise-induced phosphorylation of the novel Akt substrates AS160 and filamin A in human skeletal muscle. Diabetes. 2006;55:1776–1782. doi: 10.2337/db05-1419. [DOI] [PubMed] [Google Scholar]
- Drummond MJ, Fry CS, Glynn EL, Dreyer HC, Dhanani S, Timmerman KL, Volpi E, Rasmussen BB. Rapamycin administration in humans blocks the contraction-induced increase in skeletal muscle protein synthesis. J Physiol. 2009;587:1535–1546. doi: 10.1113/jphysiol.2008.163816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facchinetti MM, Boland R, de Boland AR. Calcitriol transmembrane signalling: regulation of rat muscle phospholipase D activity. J Lipid Res. 1998;39:197–204. [PubMed] [Google Scholar]
- Fang Y, Vilella-Bach M, Bachmann R, Flanigan A, Chen J. Phosphatidic acid-mediated mitogenic activation of mTOR signaling. Science. 2001;294:1942–1945. doi: 10.1126/science.1066015. [DOI] [PubMed] [Google Scholar]
- Fingar DC, Richardson CJ, Tee AR, Cheatham L, Tsou C, Blenis J. mTOR controls cell cycle progression through its cell growth effectors S6K1 and 4E-BP1/eukaryotic translation initiation factor 4E. Mol Cell Biol. 2004;24:200–216. doi: 10.1128/MCB.24.1.200-216.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fingar DC, Salama S, Tsou C, Harlow E, Blenis J. Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev. 2002;16:1472–1487. doi: 10.1101/gad.995802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitts RH, Riley DR, Widrick JJ. Physiology of a microgravity environment invited review: microgravity and skeletal muscle. J Appl Physiol. 2000;89:823–839. doi: 10.1152/jappl.2000.89.2.823. [DOI] [PubMed] [Google Scholar]
- Foster DA. Regulation of mTOR by phosphatidic acid? Cancer Res. 2007;67:1–4. doi: 10.1158/0008-5472.CAN-06-3016. [DOI] [PubMed] [Google Scholar]
- Frost RA, Lang CH. Protein kinase B/Akt: a nexus of growth factor and cytokine signaling in determining muscle mass. J Appl Physiol. 2007;103:378–387. doi: 10.1152/japplphysiol.00089.2007. [DOI] [PubMed] [Google Scholar]
- Fujii N, Jessen N, Goodyear LJ. AMP-activated protein kinase and the regulation of glucose transport. Am J Physiol Endocrinol Metab. 2006;291:E867–877. doi: 10.1152/ajpendo.00207.2006. [DOI] [PubMed] [Google Scholar]
- Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol. 2004;5:827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AL. Protein synthesis during work-induced growth of skeletal muscle. J Cell Biol. 1968;36:653–658. doi: 10.1083/jcb.36.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AL, Etlinger JD, Goldspink DF, Jablecki C. Mechanism of work-induced hypertrophy of skeletal muscle. Med Sci Sports. 1975;7:185–198. [PubMed] [Google Scholar]
- Goldspink DF. The influence of immobilization and stretch on protein turnover of rat skeletal muscle. J Physiol. 1977;264:267–282. doi: 10.1113/jphysiol.1977.sp011667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman CA, Mabrey DM, Frey JW, Miu MH, Schmidt EK, Pierre P, Hornberger TA. Novel insights into the regulation of skeletal muscle protein synthesis as revealed by a new nonradioactive in vivo technique. FASEB J. 2011;25:1028–1039. doi: 10.1096/fj.10-168799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman CA, Miu MH, Frey JW, Mabrey DM, Lincoln HC, Ge Y, Chen J, Hornberger TA. A phosphatidylinositol 3-kinase/protein kinase B-independent activation of mammalian target of rapamycin signaling is sufficient to induce skeletal muscle hypertrophy. Mol Biol Cell. 2010;21:3258–3268. doi: 10.1091/mbc.E10-05-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin DA, Stevens DM, Thoreen CC, Burds AA, Kalaany NY, Moffat J, Brown M, Fitzgerald KJ, Sabatini DM. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev Cell. 2006;11:859–871. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Gulati P, Gaspers LD, Dann SG, Joaquin M, Nobukuni T, Natt F, Kozma SC, Thomas AP, Thomas G. Amino acids activate mTOR complex 1 via Ca2+/CaM signaling to hVps34. Cell Metab. 2008;7:456–465. doi: 10.1016/j.cmet.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulati P, Thomas G. Nutrient sensing in the mTOR/S6K1 signalling pathway. Biochem Soc Trans. 2007;35:236–238. doi: 10.1042/BST0350236. [DOI] [PubMed] [Google Scholar]
- Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha SH, Kim DH, Kim IS, Kim JH, Lee MN, Lee HJ, Kim JH, Jang SK, Suh PG, Ryu SH. PLD2 forms a functional complex with mTOR/raptor to transduce mitogenic signals. Cell Signal. 2006;18:2283–2291. doi: 10.1016/j.cellsig.2006.05.021. [DOI] [PubMed] [Google Scholar]
- Haddad F, Adams GR. Aging-sensitive cellular and molecular mechanisms associated with skeletal muscle hypertrophy. J Appl Physiol. 2006;100:1188–1203. doi: 10.1152/japplphysiol.01227.2005. [DOI] [PubMed] [Google Scholar]
- Hahn-Windgassen A, Nogueira V, Chen CC, Skeen JE, Sonenberg N, Hay N. Akt activates the mammalian target of rapamycin by regulating cellular ATP level and AMPK activity. J Biol Chem. 2005;280:32081–32089. doi: 10.1074/jbc.M502876200. [DOI] [PubMed] [Google Scholar]
- Haq S, Kilter H, Michael A, Tao J, O’Leary E, Sun XM, Walters B, Bhattacharya K, Chen X, Cui L, Andreucci M, Rosenzweig A, Guerrero JL, Patten R, Liao R, Molkentin J, Picard M, Bonventre JV, Force T. Deletion of cytosolic phospholipase A2 promotes striated muscle growth. Nat Med. 2003;9:944–951. doi: 10.1038/nm891. [DOI] [PubMed] [Google Scholar]
- Harris TE, Huffman TA, Chi A, Shabanowitz J, Hunt DF, Kumar A, Lawrence JC., Jr. Insulin controls subcellular localization and multisite phosphorylation of the phosphatidic acid phosphatase, lipin 1. J Biol Chem. 2007;282:277–286. doi: 10.1074/jbc.M609537200. [DOI] [PubMed] [Google Scholar]
- Hooks SB, Ragan SP, Lynch KR. Identification of a novel human phosphatidic acid phosphatase type 2 isoform. FEBS Lett. 1998;427:188–192. doi: 10.1016/s0014-5793(98)00421-9. [DOI] [PubMed] [Google Scholar]
- Hornberger TA, Chien S. Mechanical stimuli and nutrients regulate rapamycin-sensitive signaling through distinct mechanisms in skeletal muscle. J Cell Biochem. 2006;97:1207–1216. doi: 10.1002/jcb.20671. [DOI] [PubMed] [Google Scholar]
- Hornberger TA, Chu WK, Mak YW, Hsiung JW, Huang SA, Chien S. The role of phospholipase D and phosphatidic acid in the mechanical activation of mTOR signaling in skeletal muscle. Proc Natl Acad Sci U S A. 2006;103:4741–4746. doi: 10.1073/pnas.0600678103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornberger TA, McLoughlin TJ, Leszczynski JK, Armstrong DD, Jameson RR, Bowen PE, Hwang ES, Hou H, Moustafa ME, Carlson BA, Hatfield DL, Diamond AM, Esser KA. Selenoprotein-deficient transgenic mice exhibit enhanced exercise-induced muscle growth. J Nutr. 2003;133:3091–3097. doi: 10.1093/jn/133.10.3091. [DOI] [PubMed] [Google Scholar]
- Hornberger TA, Stuppard R, Conley KE, Fedele MJ, Fiorotto ML, Chin ER, Esser KA. Mechanical stimuli regulate rapamycin-sensitive signalling by a phosphoinositide 3-kinase-, protein kinase B- and growth factor-independent mechanism. Biochem J. 2004;380:795–804. doi: 10.1042/BJ20040274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornberger TA, Sukhija KB, Wang XR, Chien S. mTOR is the rapamycin-sensitive kinase that confers mechanically-induced phosphorylation of the hydrophobic motif site Thr(389) in p70(S6k) FEBS Lett. 2007;581:4562–4566. doi: 10.1016/j.febslet.2007.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Manning BD. The TSC1-TSC2 complex: a molecular switchboard controlling cell growth. Biochem J. 2008;412:179–190. doi: 10.1042/BJ20080281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Li Y, Xu T, Guan KL. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 2003;17:1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumiya Y, Hopkins T, Morris C, Sato K, Zeng L, Viereck J, Hamilton JA, Ouchi N, LeBrasseur NK, Walsh K. Fast/Glycolytic muscle fiber growth reduces fat mass and improves metabolic parameters in obese mice. Cell Metab. 2008;7:159–172. doi: 10.1016/j.cmet.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacinto E, Loewith R, Schmidt A, Lin S, Ruegg MA, Hall A, Hall MN. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol. 2004;6:1122–1128. doi: 10.1038/ncb1183. [DOI] [PubMed] [Google Scholar]
- Jenkins GM, Frohman MA. Phospholipase D: a lipid centric review. Cell Mol Life Sci. 2005;62:2305–2316. doi: 10.1007/s00018-005-5195-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhasz G, Hill JH, Yan Y, Sass M, Baehrecke EH, Backer JM, Neufeld TP. The class III PI(3)K Vps34 promotes autophagy and endocytosis but not TOR signaling in Drosophila. J Cell Biol. 2008;181:655–666. doi: 10.1083/jcb.200712051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai M, Wada I, Imai S, Sakane F, Kanoh H. Cloning and characterization of two human isozymes of Mg2+-independent phosphatidic acid phosphatase. J Biol Chem. 1997;272:24572–24578. doi: 10.1074/jbc.272.39.24572. [DOI] [PubMed] [Google Scholar]
- Katayama K, Kodaki T, Nagamachi Y, Yamashita S. Cloning, differential regulation and tissue distribution of alternatively spliced isoforms of ADP-ribosylation-factor-dependent phospholipase D from rat liver. Biochem J. 1998;329(Pt 3):647–652. doi: 10.1042/bj3290647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10:935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Guan KL. RAG GTPases in nutrient-mediated TOR signaling pathway. Cell Cycle. 2009;8:1014–1018. doi: 10.4161/cc.8.7.8124. [DOI] [PubMed] [Google Scholar]
- Kim J, Solis RS, Arias EB, Cartee GD. Postcontraction insulin sensitivity: relationship with contraction protocol, glycogen concentration, and 5′ AMP-activated protein kinase phosphorylation. J Appl Physiol. 2004;96:575–583. doi: 10.1152/japplphysiol.00909.2003. [DOI] [PubMed] [Google Scholar]
- Kimball SR. Interaction between the AMP-activated protein kinase and mTOR signaling pathways. Med Sci Sports Exerc. 2006;38:1958–1964. doi: 10.1249/01.mss.0000233796.16411.13. [DOI] [PubMed] [Google Scholar]
- Kimball SR, Farrell PA, Jefferson LS. Invited Review: Role of insulin in translational control of protein synthesis in skeletal muscle by amino acids or exercise. J Appl Physiol. 2002;93:1168–1180. doi: 10.1152/japplphysiol.00221.2002. [DOI] [PubMed] [Google Scholar]
- Kimball SR, Shantz LM, Horetsky RL, Jefferson LS. Leucine regulates translation of specific mRNAs in L6 myoblasts through mTOR-mediated changes in availability of eIF4E and phosphorylation of ribosomal protein S6. J Biol Chem. 1999;274:11647–11652. doi: 10.1074/jbc.274.17.11647. [DOI] [PubMed] [Google Scholar]
- Kodaki T, Yamashita S. Cloning, expression, and characterization of a novel phospholipase D complementary DNA from rat brain. J Biol Chem. 1997;272:11408–11413. doi: 10.1074/jbc.272.17.11408. [DOI] [PubMed] [Google Scholar]
- Kubica N, Bolster DR, Farrell PA, Kimball SR, Jefferson LS. Resistance exercise increases muscle protein synthesis and translation of eukaryotic initiation factor 2Bepsilon mRNA in a mammalian target of rapamycin-dependent manner. J Biol Chem. 2004 doi: 10.1074/jbc.M413732200. [DOI] [PubMed] [Google Scholar]
- Kubica N, Bolster DR, Farrell PA, Kimball SR, Jefferson LS. Resistance exercise increases muscle protein synthesis and translation of eukaryotic initiation factor 2Bepsilon mRNA in a mammalian target of rapamycin-dependent manner. J Biol Chem. 2005;280:7570–7580. doi: 10.1074/jbc.M413732200. [DOI] [PubMed] [Google Scholar]
- Lane HA, Fernandez A, Lamb NJ, Thomas G. p70s6k function is essential for G1 progression. Nature. 1993;363:170–172. doi: 10.1038/363170a0. [DOI] [PubMed] [Google Scholar]
- Lee RC, Wang Z, Heo M, Ross R, Janssen I, Heymsfield SB. Total-body skeletal muscle mass: development and cross-validation of anthropometric prediction models. Am J Clin Nutr. 2000;72:796–803. doi: 10.1093/ajcn/72.3.796. [DOI] [PubMed] [Google Scholar]
- Lockhart NC, Baar K, Mazzeo RS, Brooks SV. Activation of Akt as a potential mediator of adaptations that reduce muscle injury. Med Sci Sports Exerc. 2006;38:1058–1064. doi: 10.1249/01.mss.0000222832.43520.27. [DOI] [PubMed] [Google Scholar]
- Long X, Ortiz-Vega S, Lin Y, Avruch J. Rheb binding to mammalian target of rapamycin (mTOR) is regulated by amino acid sufficiency. J Biol Chem. 2005;280:23433–23436. doi: 10.1074/jbc.C500169200. [DOI] [PubMed] [Google Scholar]
- Lorenz MC, Heitman J. TOR mutations confer rapamycin resistance by preventing interaction with FKBP12-rapamycin. J Biol Chem. 1995;270:27531–27537. doi: 10.1074/jbc.270.46.27531. [DOI] [PubMed] [Google Scholar]
- Lu B, Jiang YJ, Zhou Y, Xu FY, Hatch GM, Choy PC. Cloning and characterization of murine 1-acyl-sn-glycerol 3-phosphate acyltransferases and their regulation by PPARalpha in murine heart. Biochem J. 2005;385:469–477. doi: 10.1042/BJ20041348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D, Bai X, Guo S, Jiang Y. The switch I region of Rheb is critical for its interaction with FKBP38. J Biol Chem. 2008 doi: 10.1074/jbc.M802356200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKenzie MG, Hamilton DL, Murray JT, Taylor PM, Baar K. mVps34 is activated following high-resistance contractions. J Physiol. 2009;587:253–260. doi: 10.1113/jphysiol.2008.159830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney SJ, Dempsey JM, Blenis J. Chapter 2 Cell Signaling in Protein Synthesis Ribosome Biogenesis and Translation Initiation and Elongation. Prog Mol Biol Transl Sci. 2009;90C:53–107. doi: 10.1016/S1877-1173(09)90002-3. [DOI] [PubMed] [Google Scholar]
- Meyuhas O. Synthesis of the translational apparatus is regulated at the translational level. Eur J Biochem. 2000;267:6321–6330. doi: 10.1046/j.1432-1327.2000.01719.x. [DOI] [PubMed] [Google Scholar]
- Murakami M, Kudo I. Phospholipase A2. J Biochem. 2002;131:285–292. doi: 10.1093/oxfordjournals.jbchem.a003101. [DOI] [PubMed] [Google Scholar]
- Musi N, Yu H, Goodyear LJ. AMP-activated protein kinase regulation and action in skeletal muscle during exercise. Biochem Soc Trans. 2003;31:191–195. doi: 10.1042/bst0310191. [DOI] [PubMed] [Google Scholar]
- Nader GA. Molecular determinants of skeletal muscle mass: getting the “AKT” together. Int J Biochem Cell Biol. 2005;37:1985–1996. doi: 10.1016/j.biocel.2005.02.026. [DOI] [PubMed] [Google Scholar]
- Nanjundan M, Possmayer F. Pulmonary phosphatidic acid phosphatase and lipid phosphate phosphohydrolase. Am J Physiol Lung Cell Mol Physiol. 2003;284:L1–23. doi: 10.1152/ajplung.00029.2002. [DOI] [PubMed] [Google Scholar]
- Nobukuni T, Joaquin M, Roccio M, Dann SG, Kim SY, Gulati P, Byfield MP, Backer JM, Natt F, Bos JL, Zwartkruis FJ, Thomas G. Amino acids mediate mTOR/raptor signaling through activation of class 3 phosphatidylinositol 3OH-kinase. Proc Natl Acad Sci U S A. 2005;102:14238–14243. doi: 10.1073/pnas.0506925102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neil TK, Duffy LR, Frey JW, Hornberger TA. The Role of Phosphoinositide 3-kinase and Phosphatidic Acid in the Regulation of mTOR following Eccentric Contractions. J Physiol. 2009 doi: 10.1113/jphysiol.2009.173609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohanna M, Sobering AK, Lapointe T, Lorenzo L, Praud C, Petroulakis E, Sonenberg N, Kelly PA, Sotiropoulos A, Pende M. Atrophy of S6K1(-/-) skeletal muscle cells reveals distinct mTOR effectors for cell cycle and size control. Nat Cell Biol. 2005;7:286–294. doi: 10.1038/ncb1231. [DOI] [PubMed] [Google Scholar]
- Oude Weernink PA, Han L, Jakobs KH, Schmidt M. Dynamic phospholipid signaling by G protein-coupled receptors. Biochim Biophys Acta. 2007;1768:888–900. doi: 10.1016/j.bbamem.2006.09.012. [DOI] [PubMed] [Google Scholar]
- Pahor M, Kritchevsky S. Research hypotheses on muscle wasting, aging, loss of function and disability. J Nutr Health Aging. 1998;2:97–100. [PubMed] [Google Scholar]
- Pallafacchina G, Calabria E, Serrano AL, Kalhovde JM, Schiaffino S. A protein kinase B-dependent and rapamycin-sensitive pathway controls skeletal muscle growth but not fiber type specification. Proc Natl Acad Sci U S A. 2002;99:9213–9218. doi: 10.1073/pnas.142166599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pende M, Um SH, Mieulet V, Sticker M, Goss VL, Mestan J, Mueller M, Fumagalli S, Kozma SC, Thomas G. S6K1(-/-)/S6K2(-/-) mice exhibit perinatal lethality and rapamycin-sensitive 5′-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway. Mol Cell Biol. 2004;24:3112–3124. doi: 10.1128/MCB.24.8.3112-3124.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor DN, Balagopal P, Nair KS. Age-related sarcopenia in humans is associated with reduced synthetic rates of specific muscle proteins. J Nutr. 1998;128:351S–355S. doi: 10.1093/jn/128.2.351S. [DOI] [PubMed] [Google Scholar]
- Proud CG. Signalling to translation: how signal transduction pathways control the protein synthetic machinery. Biochem J. 2007;403:217–234. doi: 10.1042/BJ20070024. [DOI] [PubMed] [Google Scholar]
- Reiling JH, Sabatini DM. Stress and mTORture signaling. Oncogene. 2006;25:6373–6383. doi: 10.1038/sj.onc.1209889. [DOI] [PubMed] [Google Scholar]
- Rommel C, Bodine SC, Clarke BA, Rossman R, Nunez L, Stitt TN, Yancopoulos GD, Glass DJ. Mediation of IGF-1-induced skeletal myotube hypertrophy by PI(3)K/Akt/mTOR and PI(3)K/Akt/GSK3 pathways. Nat Cell Biol. 2001;3:1009–1013. doi: 10.1038/ncb1101-1009. [DOI] [PubMed] [Google Scholar]
- Sakamoto K, Aschenbach WG, Hirshman MF, Goodyear LJ. Akt signaling in skeletal muscle: regulation by exercise and passive stretch. Am J Physiol Endocrinol Metab. 2003;285:E1081–1088. doi: 10.1152/ajpendo.00228.2003. [DOI] [PubMed] [Google Scholar]
- Sakane F, Imai S, Kai M, Yasuda S, Kanoh H. Diacylglycerol kinases: why so many of them? Biochim Biophys Acta. 2007;1771:793–806. doi: 10.1016/j.bbalip.2007.04.006. [DOI] [PubMed] [Google Scholar]
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandri M. Signaling in muscle atrophy and hypertrophy. Physiology (Bethesda) 2008;23:160–170. doi: 10.1152/physiol.00041.2007. [DOI] [PubMed] [Google Scholar]
- Seguin R, Nelson ME. The benefits of strength training for older adults. Am J Prev Med. 2003;25:141–149. doi: 10.1016/s0749-3797(03)00177-6. [DOI] [PubMed] [Google Scholar]
- Shen Y, Xu L, Foster DA. Role for phospholipase D in receptor-mediated endocytosis. Mol Cell Biol. 2001;21:595–602. doi: 10.1128/MCB.21.2.595-602.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima H, Pende M, Chen Y, Fumagalli S, Thomas G, Kozma SC. Disruption of the p70(s6k)/p85(s6k) gene reveals a small mouse phenotype and a new functional S6 kinase. Embo J. 1998;17:6649–6659. doi: 10.1093/emboj/17.22.6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangenburg EE, Le Roith D, Ward CW, Bodine SC. A functional insulin-like growth factor receptor is not necessary for load-induced skeletal muscle hypertrophy. J Physiol. 2008;586:283–291. doi: 10.1113/jphysiol.2007.141507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangenburg EE, McBride TA. Inhibition of stretch-activated channels during eccentric muscle contraction attenuates p70S6K activation. J Appl Physiol. 2006;100:129–135. doi: 10.1152/japplphysiol.00619.2005. [DOI] [PubMed] [Google Scholar]
- Su W, Yeku O, Olepu S, Genna A, Park JS, Ren H, Du G, Gelb MH, Morris AJ, Frohman MA. 5-Fluoro-2-indolyl des-chlorohalopemide (FIPI), a phospholipase D pharmacological inhibitor that alters cell spreading and inhibits chemotaxis. Mol Pharmacol. 2009;75:437–446. doi: 10.1124/mol.108.053298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Fang Y, Yoon MS, Zhang C, Roccio M, Zwartkruis FJ, Armstrong M, Brown HA, Chen J. Phospholipase D1 is an effector of Rheb in the mTOR pathway. Proc Natl Acad Sci U S A. 2008;105:8286–8291. doi: 10.1073/pnas.0712268105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N, Nagamine M, Tanno S, Motomura W, Kohgo Y, Okumura T. A diacylglycerol kinase inhibitor, R59022, stimulates glucose transport through a MKK3/6-p38 signaling pathway in skeletal muscle cells. Biochem Biophys Res Commun. 2007;360:244–250. doi: 10.1016/j.bbrc.2007.06.040. [DOI] [PubMed] [Google Scholar]
- Tang W, Yuan J, Chen X, Gu X, Luo K, Li J, Wan B, Wang Y, Yu L. Identification of a novel human lysophosphatidic acid acyltransferase, LPAAT-theta, which activates mTOR pathway. J Biochem Mol Biol. 2006;39:626–635. doi: 10.5483/bmbrep.2006.39.5.626. [DOI] [PubMed] [Google Scholar]
- Topham MK. Signaling roles of diacylglycerol kinases. J Cell Biochem. 2006;97:474–484. doi: 10.1002/jcb.20704. [DOI] [PubMed] [Google Scholar]
- Vandenburgh H, Chromiak J, Shansky J, Del Tatto M, Lemaire J. Space travel directly induces skeletal muscle atrophy. FASEB J. 1999;13:1031–1038. doi: 10.1096/fasebj.13.9.1031. [DOI] [PubMed] [Google Scholar]
- Vandenburgh HH. Motion into mass: how does tension stimulate muscle growth? Med Sci Sports Exerc. 1987;19:S142–149. [PubMed] [Google Scholar]
- Vandenburgh HH, Kaufman S. Stretch-induced growth of skeletal myotubes correlates with activation of the sodium pump. J Cell Physiol. 1981;109:205–214. doi: 10.1002/jcp.1041090203. [DOI] [PubMed] [Google Scholar]
- Varma S, Khandelwal RL. Effects of rapamycin on cell proliferation and phosphorylation of mTOR and p70(S6K) in HepG2 and HepG2 cells overexpressing constitutively active Akt/PKB. Biochim Biophys Acta. 2007;1770:71–78. doi: 10.1016/j.bbagen.2006.07.016. [DOI] [PubMed] [Google Scholar]
- Veverka V, Crabbe T, Bird I, Lennie G, Muskett FW, Taylor RJ, Carr MD. Structural characterization of the interaction of mTOR with phosphatidic acid and a novel class of inhibitor: compelling evidence for a central role of the FRB domain in small molecule-mediated regulation of mTOR. Oncogene. 2008;27:585–595. doi: 10.1038/sj.onc.1210693. [DOI] [PubMed] [Google Scholar]
- Wakelam MJ, Hodgkin M, Martin A. The measurement of phospholipase D-linked signaling in cells. Methods Mol Biol. 1995;41:271–278. doi: 10.1385/0-89603-298-1:271. [DOI] [PubMed] [Google Scholar]
- Wang X, Devaiah SP, Zhang W, Welti R. Signaling functions of phosphatidic acid. Prog Lipid Res. 2006 doi: 10.1016/j.plipres.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Winter JN, Fox TE, Kester M, Jefferson LS, Kimball SR. Phosphatidic acid mediates activation of mTORC1 through the ERK signaling pathway. Am J Physiol Cell Physiol. 299:C335–344. doi: 10.1152/ajpcell.00039.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Flinn RJ, Wu H, Schnur RS, Backer JM. hVps15, but not Ca2+/CaM, is required for the activity and regulation of hVps34 in mammalian cells. Biochem J. 2009;417:747–755. doi: 10.1042/BJ20081865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Guan KL. Expanding mTOR signaling. Cell Res. 2007;17:666–681. doi: 10.1038/cr.2007.64. [DOI] [PubMed] [Google Scholar]