Abstract
Transcription factors direct gene expression, so there is much interest in mapping their genome-wide binding locations. Current methods do not allow for the multiplexed analysis of TF binding, and this limits their throughput. We describe a novel method for determining the genomic target genes of multiple transcription factors simultaneously. DNA-binding proteins are endowed with the ability to direct transposon insertions into the genome near to where they bind. The transposon becomes a “Calling Card” marking the visit of the DNA-binding protein to that location. A unique sequence “barcode” in the transposon matches it to the DNA-binding protein that directed its insertion. The sequences of the DNA flanking the transposon (which reveal where in the genome the transposon landed) and the barcode within the transposon (which identifies the TF that put it there) are determined by massively parallel DNA sequencing. To demonstrate the method's feasibility, we determined the genomic targets of eight transcription factors in a single experiment. The Calling Card method promises to significantly reduce the cost and labor needed to determine the genomic targets of many transcription factors in different environmental conditions and genetic backgrounds.
Transcription factors (TFs) regulate gene expression in response to environmental changes and developmental signals. Identification of the target genes of TFs under different conditions is essential for an understanding of transcriptional regulation. While it is straightforward to determine the recognition sequence of a TF in vitro, its in vivo targets cannot be accurately predicted from that information because not all potential TF binding sites are actually bound by the TF (Liu et al. 2005, 2006). Thus, the in vivo gene targets of a TF must be experimentally determined. Chromatin immunoprecipitation coupled with DNA microarrays (ChIP-chip) has been used to map the global in vivo binding patterns of many transcription factors of Saccharomyces cerevisiae (Harbison et al. 2004). With the help of phylogenetic alignment, recognition sequences (motifs) were identified for 98 of these transcription factors (Harbison et al. 2004; MacIsaac et al. 2006), demonstrating the power of ChIP-based methods. However, these methods lack the throughput to analyze the complete set of yeast TFs under more than just a few environmental conditions (Harbison et al. 2004), which could account for their failure to identify target genes and recognition sequences for more than half of the TFs analyzed.
Here we describe Calling Card–seq, a novel high-throughput method for determining the genomic targets of multiple transcription factors simultaneously, and use it to map the targets of eight TFs in a single experiment. The Calling Card method involves fusing to the TF a piece of the Sir4 protein that physically interacts with the Ty5 integrase. This chimeric protein recruits the Ty5 integrase, which directs integration of a Ty5 transposon into the genome near to where the TF is bound (Fig. 1A; Zhu et al. 2003; Wang et al. 2007). By “barcoding” transposons with sequence identifiers matched to each DNA-binding protein, every Calling Card is marked with a signature that indicates which protein deposited it into the genome. Transposon Calling Cards are harvested from genomic DNA by digestion with restriction endonucleases followed by circularization of the resulting fragments and their amplification in an inverse PCR with primers complementary to the transposon sequence. The DNA sequence of the genomic region immediately flanking the Calling Card (which reveals where in the genome the Calling Card landed) and the DNA sequence of the barcode (which reveals which TF was responsible for depositing the Calling Card there) are determined by paired-end DNA sequencing on an Illumina GAII instrument. This approach enables simultaneous analysis of multiple DNA-binding proteins (Fig. 1B).
Figure 1.
Calling Card–seq. (A) A DNA-binding protein fused to Sir4 directs integration of Ty5 into the genome near to where it binds. (1) After Ty5 transposition, (2) cells that have undergone Ty5 transposition are selected. (3) Genomic DNA is isolated and cleaved with restriction enzymes that cut near the end of Ty5 and (4) ligated in a dilute solution to favor recircularization of the fragments. (5) This is followed by amplification of the circular DNA that contains the end of the transposon and flanking genomic DNA by an “inverse PCR” (the PCR primers contain the Illumina sequencing primers and adaptors). The DNA sequence of the inverse-PCR products is then determined on an Illumina Genome Analyzer II. (B) Analyzing multiple TFs in one experiment: (1) Each strain was cotransformed with a plasmid encoding a TF–Sir4 fusion and a plasmid carrying its matched barcoded Ty5 Calling Card. (2) After transposition, the Calling Cards are deposited across the genome and then (3) recovered by inverse PCR and (4) sequenced on the Illumina GAII with a paired-end module. (5) For each paired sequence, we identify the Calling Card insertion site and the TF that deposited it there.
We believe that the parallel nature and ease of use of Calling Card–seq make it an attractive approach for the identification or validation of transcription factor targets that is orthogonal to ChIP-based methods; it should be especially useful for analyzing large sets of TFs under many different conditions or in many genetic backgrounds.
Results
Calling Card–seq accurately maps transcription factor binding in vivo
To validate the Calling Card–seq method, we applied it to three well-studied TFs: Gal4, Gcn4, and Leu3. For each TF, we mapped more than 5000 independent Ty5 insertions. The global patterns of Ty5 insertions were dramatically different in the strains with Sir4 fused to one of these TFs compared to a control strain that lacked a TF–Sir4 fusion (Fig. 2A), suggesting that the majority of transposon insertions were directed by TF binding. This was further supported by the observation that Calling Cards were specifically deposited near known binding sites for these TFs (Fig. 2B).
Figure 2.
Calling Card–seq accurately predicts target genes and DNA-binding motifs. (A) The genome-wide Ty5 insertion patterns of Gal4, Leu3, Gcn4, and no-TF control. (B) Ty5 integrations are enriched around Gal4 binding sites (indicated by the cyan triangles) in the GCY1 promoter. The x-axis specifies gene position; the y-axis is the number of sequencing reads for each insertion (indicated by the blue circle). Each blue circle represents a Calling Card deposited at a unique location. (C) ROC curves for Gal4 (red), Leu3 (green), and Gcn4 (blue). (D) The distribution of Gcn4-directed Ty5 insertions around known Gcn4p-binding sites. The x-axis specifies the distance from the center of the Gcn4-binding site; the y-axis is the number of insertion events.
We developed a quantitative method for identifying the genomic targets of a TF from Calling Card–seq data. Using data obtained from the “no TF–Sir4” control strain, we constructed a statistical model that describes the natural tendency of the Ty5 integrase to deposit Calling Cards into each promoter in the yeast genome (see Methods). This model allowed us to compute whether the observed number of transposition events in a given promoter is greater than predicted by chance. The stringency with which target genes are identified can be adjusted by using different probability (P-value) cutoffs (Supplemental Table 1).
Receiver–operator curves (Lusted 1971), which are plots of the sensitivity of the method (how many known targets are identified) versus the false positive rate (or 1 − specificity) at different statistical cutoffs, reveal the sensitivity and specificity of Calling Card–seq (Fig. 2C). The area under a receiver–operator curve (AUC) provides a measure of the accuracy of the method. An area of 1 indicates that the method is perfectly accurate; an area of 0.5 indicates that the method is performing as expected by chance. The AUC for the Calling Card–seq method is 0.99, 0.84, and 0.99 for Gal4, Gcn4, and Leu3, respectively (Fig. 2C), demonstrating that the method is quite accurate (few false positives). For example, 100% of the known Leu3 targets were identified at a false-positive rate of 1.3%.
Within the promoters of target genes of these TFs, we observed that Calling Card insertions were highly enriched around TF binding sites (Fig. 2B). Since we determine the locations of Ty5 transposons with single-nucleotide resolution, we can calculate the distribution of Ty5 insertions around known protein binding sites with high precision. A plot of the frequency of Calling Cards deposited by Gcn4 as a function of distance from known Gcn4-binding sites (Fig. 2D) reveals that most Gcn4-directed insertions (>60%) occurred within 100 bp of the Gcn4-binding site (50 bp on each side of the site). Conversely, most Gcn4-binding sites were located close to the center of a Calling Card cluster. The median distance from the peak center to the nearest Gcn4-binding site was 9 bp (standard deviation = 72 bp). Similar patterns were observed for Gal4 and Leu3 (Supplemental Fig. 1). There were strikingly few insertions directly into the binding site (note the sharp dip in the histogram from −5 to +5 bases in Fig. 2D), presumably because the transcription factor sterically blocks integration at those nucleotides. The tight clustering of insertion events around binding sites for a transcription factor facilitates the identification of the sequence motif it recognizes because it limits the sequence search space (Kharchenko et al. 2008), making it relatively straightforward to infer the position-specific weight matrix (PSWM) of a transcription factor using Calling Card data. We searched for a PSWM for each TF by analyzing with the AlignACE algorithm (see Methods) the DNA sequence in the region of the genome where Calling Cards were inserted (Roth et al. 1998). Previously known motifs for all three TFs were successfully identified (Supplemental Fig. 2). We conclude that the Calling Card method can be used to determine the recognition sequences of transcription factors, in addition to identifying in vivo gene targets.
Calling Card–seq enables analysis of multiple TFs in a single experiment
Analysis of TFs by Calling Card–seq can be multiplexed if unique sequence identifiers (“barcodes”) are included in the Ty5 transposon. We tested this capability with seven TFs whose consensus recognition sequence motifs were not revealed by ChIP-chip experiments (Harbison et al. 2004; MacIsaac et al. 2006), with Gal4 included as a positive control. Eight yeast strains, each carrying a different TF fused to Sir4 and a Ty5 transposon carrying a unique 5-bp “barcode,” were pooled, and the TFs were allowed to deposit their Calling Cards (see Methods). Two “paired-end” sequencing reads were obtained for each recovered Calling Card: The first identifies the genomic sequence immediately flanking the Calling Card; the second yields the unique sequence “barcode” that reveals the TF responsible for depositing the Calling Card (Fig. 1B). More than 6 million paired-end DNA sequence reads were obtained, 62% of which contained both the barcode sequence and a genomic sequence that maps uniquely in the yeast genome. For each of the eight TFs, we were able to map more than 4500 independent insertions (Supplemental Table 2). All previously known Gal4 target genes were identified, indicating that the multiplexed method is accurate. It was also highly reproducible (Fig. 3A; Supplemental Fig. 3).
Figure 3.
Multiplexing experiments are reproducible and productive, and the Calling Card–seq method performs well when the TF–Sir4 is expressed from its native genomic locus. (A) The number of independent Calling Card insertions within each promoter is plotted for two biological replicate experiments multiplexing eight TFs. Data for Yrm1 and Sef1 are shown here. (B) Sequence logos for newly discovered TF binding site motifs.
We were able to predict recognition sequence motifs for three TFs (Yrm1, Rgm1, and Sef1) (Fig. 3B). The PSWM we predicted for Yrm1 is nearly identical to that predicted from studies that employed protein binding microarrays (Badis et al. 2008; Zhu et al. 2009). We predicted AGGGGNGGGG as the sequence recognized by Rgm1 (Fig. 3B). Badis et al. (2008) predicted CAGGGG, suggesting they identified the half-site for this protein. (Zhu et al. 2009 were unable to identify a recognition motif for Rgm1.) Similar discrepancies between a TF's in vitro and in vivo binding preferences have been previously observed (Haribson et al. 2004; MacIsaac et al. 2006; Badis et al. 2008). The recognition motif that we predicted for Sef1 has not been previously reported (Fig. 3B). We verified all three of these newly identified motifs in a bacterial one-hybrid assay (Supplemental Fig. 4). For Kar4 and Rpi1, we were unable to identify a motif with high information content, despite the fact that we could identify their target genes reproducibly (Supplemental Fig. 3; Supplemental Tables 3, 4). These results indicate that Kar4 and Rpi1 may bind a PSWM with low information content missed by our search and may require coactivators for their specificity. Consistent with this hypothesis, previous work suggests that Kar4 requires Ste12 to bind to its targets in vivo and in vitro (Lahav et al. 2007), and we were able to identify a weak Ste12 motif upstream of Kar4 target genes. The patterns of Calling Card insertions deposited by Lee1 and Sfg1 are similar to a control strain that lacked a TF–Sir4 fusion, which suggests that, if the Sir4 has not disrupted their DNA-binding specificity, these proteins may not bind to DNA, or may bind to DNA nonspecifically (Supplemental Fig. 5) and may not be bona fide transcription factors.
Our lists of target genes for the transcription factors analyzed are congruent with the known biological functions of these transcription factors, suggesting that we are predicting relevant target genes. Sixteen of the 23 Yrm1 targets predicted by Lucau-Danila et al. (2003) from expression profiling and in vivo chromatin IP experiments are found in our target gene list for Yrm1 (Supplemental Tables 3, 4). Furthermore, the predicted targets are enriched for drug transmembrane transporter activity (GO:0015238, P = 3.20×10−4) as determined by an analysis of Gene Ontology (GO) terms (AmiGO ver1.7) (Ashburner et al. 2000), suggesting that they are true targets of Yrm1, a transcription factor known to be involved in multidrug resistance. For Sef1, Rgm1, and Kar4, we also observed a statistically significant enrichment of GO terms in their target gene lists. The Kar4 target list is highly enriched in genes involved in sexual reproduction (GO:0019953, P = 4.06×10−12) and mating projection (GO:0005937, P = 1.37×10−6), consistent with Kar4's known function in the pheromone response pathway (Kurihara et al. 1996; Lahav et al. 2007). Taken together, these results demonstrate that Calling Card–seq can analyze multiple transcription factors in parallel and is accurate, high-throughput, and can be used to discover novel target genes and binding motifs for transcription factors.
Calling Card–seq is functional when TF–Sir4 is expressed from native genomic locus
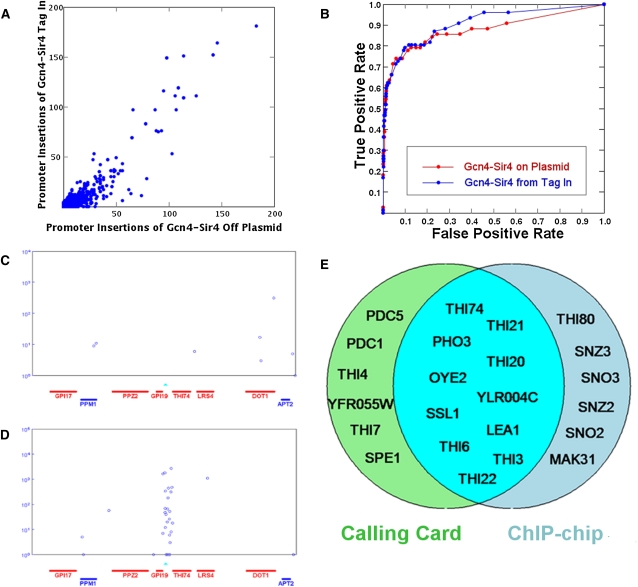
The experiments described thus far employed TF–Sir4 fusion proteins expressed from the ADH1 promoter on a plasmid. It may be preferable to express the TF from its native promoter in the genome, and when this was done for Gcn4, we observed a pattern of Calling Card deposition similar to that obtained when the Gcn4–Sir4 fusion protein was expressed from a plasmid (Fig. 4A,B), suggesting that native levels of expression are sufficient for the Calling Card method.
Figure 4.
The Calling Card–seq method performs well when the TF–Sir4 is expressed from its native genomic locus. (A) Gcn4–Sir4 fusion expressed from the ADH1 promoter on a plasmid and from its native promoter in the genome produced highly correlated Calling Card insertions. (B) ROC curves are plotted for the Calling Card data when Gcn4 is expressed from the ADH1 promoter on a plasmid (red) and from its native promoter in the genome (blue). (C) Thi2-directed Calling Cards are not enriched in the promoter of THI74 when cells are grown in thiamine-containing media. (D) Thi2-directed Calling Cards are enriched in the promoter of THI74 in cells starved for thiamine. The x-axis specifies gene position; the y-axis is the number of sequencing reads for each insertion (indicated by the blue circle). Each blue circle represents a Calling Card deposited at a unique location. Known Thi2 binding sites are indicated by the cyan triangles. (E) The target genes identified by Calling Card and ChIP methods overlap significantly.
To test whether the TF–Sir4 fusion expressed from its native promoter is responsive to environmental changes, we determined the locations of Calling Cards deposited by Thi2–Sir4 in cells grown with and without thiamine. There were substantial differences in Thi2 binding in the two conditions (Fig. 4C,D). In cells supplemented with thiamine, Thi2 showed little specific binding, with no promoter showing a P-value below 1.0×10−2 (Supplemental Table 5). In cells starved for thiamine, the promoters of 17 genes showed significant numbers of Calling Cards, and these genes are highly enriched for those involved in thiamine biosynthesis (GO:0009228, P = 9.40×10−11). These 17 genes overlapped considerably (hypergeometric P = 4.98×10−26) with those identified as Thi2 targets by ChIP-chip (Fig. 4E; Harbison et al. 2004). A smaller number of genes, most known to be involved in thiamin synthesis or regulated by thiamine, were revealed as Thi2 targets by only one of the methods, highlighting the importance of Calling Cards as an orthogonal approach to ChIP.
Discussion
We have described an easy, effective, and economical method for mapping genomic targets of DNA-binding proteins. The ability to simultaneously analyze multiple transcription factors should enable a systematic exploration of transcription factor binding in many different environments and genetic backgrounds in a way that has heretofore not been possible. The TF target genes we identified provide clues to the functions of these poorly characterized TFs.
The false-positive and false-negative rates of identifying transcription factor target genes using Calling Cards can be gleaned from the ROC curves shown in Figure 2, which are a graphical representation of the true-positive rate plotted against the false-positive rate at different P-value thresholds. False-positive and false-negative events were identified by comparing our data to ChIP-chip, which was used as the “gold standard” for this analysis. In calling targets of the seven poorly characterized TFs, we conservatively defined target genes as those whose promoters had a P-value <0.001. Based on the positive controls, this cutoff ensures a high sensitivity for TFs with a moderate number of target genes (e.g., 75% for Gal4 and 100% for Leu3), but lower sensitivity for TFs with a large number of target genes (e.g., 49.35% for Gcn4), while maintaining a high level of specificity (few false positives): 98.22% for Gal4, 98.67% for Leu3, and 99.00% for Gcn4.
Ty5 is a retrotransposon, thus once a Calling Card is inserted into a genomic locus, it cannot be removed. Therefore, Calling Cards permanently record transcription factor binding events, allowing transcription factor binding to be recorded throughout developmental processes. For example, by recording Ste12 binding during sporulation, it will be possible to compare the transcription factor's gene targets in cells that undergo sporulation with those in the cells that do not ultimately sporulate. Analysis of other dynamic processes such as filamentous growth, cellular aging, and the cell cycle would also benefit from this useful feature of the Calling Card method. We have ported the Calling Card method to mouse, human, and zebrafish cells (H Wang, D Mayhew, X Chen, M Johnston, R Mitra, in prep.), thus this method may also be applied to the study of developmental programs in multicellular organisms.
A potential limitation of this method is that Calling Cards may inactivate some genes when deposited into their promoters, preventing the analysis of TF binding at these genes. We believe that is unlikely because (1) we use diploid cells for our experiments, and (2) it is likely that some transposon insertions in a promoter will not abolish promoter function (Johnston and Davis 1984). Indeed, we found that Calling Cards were deposited into promoters of essential genes at the same frequency as they were deposited into promoters of nonessential genes in our “no TF–Sir4” control experiments (average number of insertions per essential gene promoter = 0.83 ± 1.8; average number of insertions per nonessential gene promoter = 0.91 ± 2.0; total number of insertions = 6671; P-value = 0.18). In addition, Calling Cards deposited by two well-characterized transcription factors distribute equally in essential and nonessential genes: All five essential gene targets and all eight nonessential gene targets of Leu3 received Calling Cards; three of seven (43%) essential gene targets and 38 of 70 (54%) nonessential gene targets of Gcn4 received Calling Cards. We conclude that there is no restriction in the types of genes from which Calling Cards can be recovered.
The Calling Card–seq method is easily implemented. The protocol employs general techniques of molecular biology, such as DNA cloning, restriction endonuclease digestion, DNA ligation, and PCR. The method is also flexible. It can be used to perform the multiplexed analysis of many TFs, as reported here, or it can be used to map the genome-wide DNA-binding patterns of one TF in many different mutant strains in a single experiment by “barcoding” each strain. Finally, Calling Card–seq is cost-effective: Calling Cards deposited by 10 to 20 yeast TFs can be identified on a single lane of an Illumina GAII flowcell.
Multiplexing of the Calling Card–seq method is simpler and more economical than a multiplexed ChIP-seq experiment, because all TF strains being tested are pooled at the beginning of the experiment. Testing multiple TFs by ChIP-seq requires growth of separate cultures, separate immunoprecipitations, and separate barcoded library preparations before being pooled together for DNA sequencing. With Calling Card–seq, each strain carrying a Sir4-tagged TF and a uniquely barcoded Ty5 is pooled together. One culture is grown; one DNA isolation, digestion, ligation, and inverse PCR (analogous to the immunoprecipitation for the Chip-seq experiment) is performed to create the barcoded sequencing library. In effect, implementing Calling Card–seq in this way enables one to perform eight ChIP-seq experiments in a single experiment. There is some up-front labor required to make strains containing the Sir4-tagged TFs. However, this requirement could be avoided if a library of all TF fusions is made available to the community (as was done with the yeast deletion collection).
Our goal is to multiplex up to 200 TFs of yeast, each as a TF–Sir4 expressed from its native genomic locus, and test many different growth conditions. To be able to achieve this end and to ensure adequate representation of each TF–Sir4 strain in the pool, we will need to make modest improvements in Ty5 transposition efficiency to provide higher throughput, and we will need to shorten the time necessary for induction of Ty5 transposition. Application of our method promises to bring us closer to the goal of having a complete list of target genes and sequence recognition motifs of all yeast TFs under many different conditions and in many genetic backgrounds. Ultimately, this will help us to understand better the relationship among environmental signals, TF binding, epigenetic status, and gene expression regulation in a systematic way.
Methods
Strains and media
All Calling Card experiments used diploid sir4Δ yeast strain YM7635: MATa/MATα his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 ura3Δ0/ura3Δ0 met15Δ0/MET15 LYS2/lys2Δ0 sir4∷KanMX/sir4::KanMX trp1∷Hyg/trp1∷Hyg, except for the experiment with Gcn4–Sir4 expressed from the GCN4 promoter, which used the haploid strain YM7691: MATa his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 sir4∷KanMX trp1∷Hyg GCN4∷sir4, and the experiment with Thi2–Sir4 expressed from the THI2 promoter, which used the haploid strain MATa his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 sir4∷KanMX trp1∷Hyg THI2∷sir4. Yeasts were grown in complete synthetic media with the addition of 2% glucose or galactose.
Construction of plasmids
All TF–Sir4 fusion constructs were derived from plasmid pBM5037 (Gal4DBD-Sir4-Myc) (Wang et al. 2007). The entire ORF of each TF was amplified in a PCR and used to replace Gal4DBD by homologous recombination (“gap repair”) by its cotransformation of yeast cells with the plasmid linearized by cleavage with XhoI (it cuts once in the Gal4DBD coding sequence) (Ma et al. 1987; Wach et al. 1994).
Ty5 donor plasmid pBM5249 is derived from plasmid pBM5218 (Wang et al. 2008) (it carries the Ty5 transposon with URA3 as the selectable marker). The HIS3AI marker within Ty5 was converted to HIS3. A 34-bp sequence containing partial Illumina sequencing primer 2; 5-bp barcode 1; and Hinp1I, HpaII, and TaqI recognition sequences was cloned between the FseI and PacI sites located between the 3′ LTR and the HIS3 gene within Ty5. All other barcoded Calling Cards were derived from pBM5249.
Induction of Ty5 transposition and inverse PCR
For multiplexing experiments, each strain carrying one TF–Sir4 construct and a uniquely barcoded Calling Card was grown to saturation individually in 5 mL of Glu −Trp −His media. Cultures of all eight strains were pooled and plated on 50 Gal −Trp −His plates and incubated for 2 d at room temperature to induce Ty5 transposition. After induction of transposition, cells were replica-plated to YPD media and grown for 1 d to allow them to lose the Ty5 donor plasmid. Cells were then serially replica-plated onto −His, FOA-containing media twice to select for cells containing Calling Cards in their genome.
To map the locations of the Calling Cards in the genome, all His+ FOAr colonies were harvested, and their genomic DNA was extracted. Each DNA sample was divided into three aliquots and digested with Hinp1I or HpaII or TaqI. Digested DNA was ligated overnight at 15°C in dilute solution (<10 ng/μL) to encourage self-circularization. After ethanol precipitation, self-ligated DNA was resuspended in ddH2O and used as template in an inverse PCR. Primers that anneal to Ty5 sequences (OM8714: AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTAATTCACTACGTCAACA; OM8827: CAAGCAGAAGACGGCATACGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATC) were used to amplify the genomic regions flanking Ty5 integrations and the barcodes within Ty5, as well as adding adapter sequences that allow the PCR products to be sequenced on the Illumina GA analyzer. The PCR products were purified using the QIAquick PCR Purification Kit (QIAGEN) and diluted to 10 nM. For each sample, the same amount of PCR product from digestion with each restriction endonuclease was pooled and submitted for sequencing on the Illumina GAII.
Paired-end sequence map back
DNA sequence reads were filtered by requiring the correct 17-bp LTR sequence in the first 17 bases of sequence from the first paired-end read, and an appropriate 8-bp barcode sequence and restriction enzyme digestion site sequence on the second paired-end read. Paired reads were mapped using a seeded hash approach. The first 12 genomic bases of each read were mapped to the yeast genome. The remaining genomic fragments of each read were then aligned to the seeds using an ungapped alignment and allowing up to two mismatches on each read. All alignments from the first read are compared to all alignments from the second read and screened for three requirements: (1) same chromosome, (2) within 1200 bp, and (3) opposite strands. If a matched read pair passes all these tests, it is accepted as a Calling Card insertion site. If multiple matched read pairs meet these requirements, the pair with the fewest cumulative mismatches in the alignment is accepted as an insertion site. If there is a tie and multiple pairs have the same minimum number of mismatches, the read is discarded. To eliminate bleedthrough from other barcodes and misalignments from sequencing errors, a threshold of 10 reads was then required to consider an insertion as real.
Target gene calling and sequence motif finding
Promoters were defined as the 1000 bp 5′ of the transcription start site of a gene, or until the coding sequence of the next (upstream) gene, but with a minimum size of 250 bp. The background frequency of Ty5 insertions into the yeast genome in a strain carrying no TF–Sir4 was used to create a null model for each promoter. More than 80,000 of these Calling Card insertions were collected and mapped. For every transcription factor experiment, each promoter was modeled with the Poisson distribution, with the observed counts being the number of TF–Sir4-directed Calling Card insertions in that promoter, and the expected value equaling the average number of insertions in that promoter from a Monte Carlo sampling of the no TF–Sir4 insertions (based on the number of TF–Sir4 insertions collected in that experiment), plus a pseudocount. P-values were assigned at each promoter by calculating the cumulative distribution function for the promoter's observed insertions in relation to its null model-generated Poisson distribution.
We conservatively defined target genes as those whose promoters had a P-value <0.001. Based on the positive controls, this cutoff ensured a high sensitivity for TFs with a moderate number of targets (75% for Gal4 and 100% for Leu3), but lower sensitivity for TFs with a large number of targets (49.35% for Gcn4), while maintaining a high level of specificity (few false positives) (98.22% for Gal4, 98.67% for Leu3, and 99.00% for Gcn4).
To find binding motifs, we first defined “insertion frames” as the set of 100-bp sequences flanking Ty5 insertions. We concatenated overlapping insertion frames into longer contiguous sequences by requiring a minimum number of overlaps between insertion frames. The initial cutoff for number of overlapping insertion frames was determined by requiring that every promoter with a P-value <0.001 has at least one Calling Card cluster. The concatenated sequences were then searched for over-represented sequence motifs using AlignACE (Roth et al. 1998). The cutoff was adjusted both up and down to see if higher or low cutoffs would reveal a higher scoring motif. Binding potentials for the five highest information content sequence motifs from AlignACE were generated across the genome using GOMER (Granek and Clarke 2005). The motif that most accurately predicted binding as determined by area under the receiver–operator curve was selected.
Generating ROC curves for Calling Card data
Receiver–operator curves plot the sensitivity versus 1 − Specificity. To calculate sensitivity, positive lists for known TF targets were generated from the literature: Gal4 (Ren et al. 2000; Wang et al. 2007), Gcn4 (Pokholok et al. 2005), and Leu3 (Harbison et al. 2004). To calculate specificity, lists of 900 genes that are unlikely to be targets of Gal4, Gcn4, and Leu3 were randomly selected from a list of genes whose promoters (1) had a P-value >0.05 in the data set of Harbison et al. (2004); (2) were not within two genes in either direction of a strong target (P-value < 0.0001 in that data set); and (3) did not contain strong or weak binding sites for the known PWM of that transcription factor as defined by being in the lower half of all promoters as ranked by GOMER with the default Gaussian parameters.
Bacterial one-hybrid
The expression (bait) plasmid was created by amplifying the ORF of the transcription factor in a PCR and adding a Kpn1 site on the 5′ primer and a XbaI (or NheI) site on the 3′ primer and cloning the subsequent product into the Kpn1–XbaI sites of pB1H2ω5 in frame with omega (Meng et al. 2005). Positive reporter sequences were chosen by picking a genomic target containing a single site matching the consensus sequence of the predicted motif with 4–5 bp of flanking sequence on both sides. The negative reporter sequence was identical except the high information content positions in the predicted motif were altered. Both positive and negative reporter constructs were created by cloning the respective sequences into pH3U3-Zif268 using the Not1–EcoRI sites.
Both expression and reporter plasmids were cotransformed into competent Escherichia coli (with deletions in both the hisB and pyrF genes) for both the positive and negative reporters, and the concentration of the dual-transformed cells was determined by serial dilution on selective plates containing kanamycin (25 μg/mL) and carbenicillin (100 μg/mL). Equal amounts of dual-transformed cells for both positive and negative reporters were then grown on plates containing kanamycin, carbenicillin, and increasing concentrations of 3-AT ranging from 0 mM to 8 mM and grown at 37°C for 40 h.
Acknowledgments
We are grateful to Jessica Hoisington-Lopez, Laura Langton, Brian Koebbe, and Jim Dover (Washington University) for their expert assistance with the use of the Illumina Genome Analyzer. Next-generation sequencing was performed at the Washington University Genomic Technology Access Center. This work was supported by NIH grants R21RR023960, 5R01DA025744-02, R01GM078222, 5P50HG003170-03, and T32 HG000045, and funds provided by the James S. McDonnell Foundation.
Footnotes
[Supplemental material is available for this article. The sequence data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo) under accession no. GSE27381.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.114850.110.
References
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. 2000. Gene Ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badis G, Chan ET, van Bakel H, Pena-Castillo L, Tillo D, Tsui K, Carlson CD, Gossett AJ, Hasinoff MJ, Warren CL, et al. 2008. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol Cell 32: 878–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granek JA, Clarke ND 2005. Explicit equilibrium modeling of transcription-factor binding and gene regulation. Genome Biol 6: R87 doi: 10.1186/gb-2005-6-10-r87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, MacIsaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, et al. 2004. Transcriptional regulatory code of a eukaryotic genome. Nature 431: 99–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M, Davis RW 1984. Sequences that regulate the divergent GAL1–GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol 4: 1440–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Tolstorukov MY, Park PJ 2008. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotechnol 26: 1351–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara LJ, Stewart BG, Gammie AE, Rose MD 1996. Kar4p, a karyogamy-specific component of the yeast pheromone response pathway. Mol Cell Biol 16: 3990–4002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahav R, Gammie A, Tavazoie S, Rose MD 2007. Role of transcription factor Kar4 in regulating downstream events in the Saccharomyces cerevisiae pheromone response pathway. Mol Cell Biol 27: 818–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Noll DM, Lieb JD, Clarke ND 2005. DIP-chip: Rapid and accurate determination of DNA-binding specificity. Genome Res 15: 421–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Lee CK, Granek JA, Clarke ND, Lieb JD 2006. Whole-genome comparison of Leu3 binding in vitro and in vivo reveals the importance of nucleosome occupancy in target site selection. Genome Res 16: 1517–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucau-Danila A, Delaveau T, Lelandais G, Devaux F, Jacq C 2003. Competitive promoter occupancy by two yeast paralogous transcription factors controlling the multidrug resistance phenomenon. J Biol Chem 278: 52641–52650 [DOI] [PubMed] [Google Scholar]
- Lusted LB 1971. Decision-making studies in patient management. N Engl J Med 284: 416–424 [DOI] [PubMed] [Google Scholar]
- Ma H, Kunes S, Schatz PJ, Botstein D 1987. Plasmid construction by homologous recombination in yeast. Gene 58: 201–216 [DOI] [PubMed] [Google Scholar]
- MacIsaac KD, Wang T, Gordon DB, Gifford DK, Stormo GD, Fraenkel E 2006. An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinformatics 7: 113 doi: 10.1186/1471-2105-7-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Brodsky MH, Wolfe SA 2005. A bacterial one-hybrid system for determining the DNA-binding specificity of transcription factors. Nat Biotechnol 23: 988–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, et al. 2005. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122: 517–527 [DOI] [PubMed] [Google Scholar]
- Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, et al. 2000. Genome-wide location and function of DNA binding proteins. Science 290: 2306–2309 [DOI] [PubMed] [Google Scholar]
- Roth FP, Hughes JD, Estep PW, Church GM 1998. Finding DNA regulatory motifs within unaligned noncoding sequences clustered by whole-genome mRNA quantitation. Nat Biotechnol 16: 939–945 [DOI] [PubMed] [Google Scholar]
- Wach A, Brachat A, Pohlmann R, Philippsen P 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10: 1793–1808 [DOI] [PubMed] [Google Scholar]
- Wang H, Johnston M, Mitra RD 2007. Calling cards for DNA-binding proteins. Genome Res 17: 1202–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Heinz ME, Crosby SD, Johnston M, Mitra RD 2008. ‘Calling Cards’ method for high-throughput identification of targets of yeast DNA-binding proteins. Nat Protoc 3: 1569–1577 [DOI] [PubMed] [Google Scholar]
- Zhu Y, Dai J, Fuerst PG, Voytas DF 2003. Controlling integration specificity of a yeast retrotransposon. Proc Natl Acad Sci 100: 5891–5895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Byers K, McCord R, Shi Z, Berger M, Newburger D, Saulrieta K, Smith Z, Shah M, Radhakrishnan M, et al. 2009. High-resolution DNA binding specificity analysis of yeast transcription factors. Genome Res 19: 556–566 [DOI] [PMC free article] [PubMed] [Google Scholar]