Abstract
We have developed a self-inactivating PiggyBac transposon system for tamoxifen inducible insertional mutagenesis from a stably integrated chromosomal donor. This system, which we have named ‘Slingshot’, utilizes a transposon carrying elements for both gain- and loss-of-function screens in vitro. We show that the Slingshot transposon can be efficiently mobilized from a range of chromosomal loci with high inducibility and low background generating insertions that are randomly dispersed throughout the genome. Furthermore, we show that once the Slingshot transposon has been mobilized it is not remobilized producing stable clonal integrants in all daughter cells. To illustrate the efficacy of Slingshot as a screening tool we set out to identify mediators of resistance to puromycin and the chemotherapeutic drug vincristine by performing genetrap screens in mouse embryonic stem cells. From these genome-wide screens we identified multiple independent insertions in the multidrug resistance transporter genes Abcb1a/b and Abcg2 conferring resistance to drug treatment. Importantly, we also show that the Slingshot transposon system is functional in other mammalian cell lines such as human HEK293, OVCAR-3 and PE01 cells suggesting that it may be used in a range of cell culture systems. Slingshot represents a flexible and potent system for genome-wide transposon-mediated mutagenesis with many potential applications.
INTRODUCTION
The complexity of the genes, pathways and networks that dictate many cellular phenotypes rarely makes it possible to employ a candidate gene (reverse genetic) approach to identify potential mediators of a biological process. In contrast genome-wide forward genetic screens, which may be performed without making a priori assumptions about the candidature of individual genes in a process, represent a powerful approach for gene discovery. Classically forward genetic screens in higher order organisms have been performed using ionizing radiation, chemical mutagens, or viruses each of which are likely to target a different repertoire of genes. While these approaches can be extremely efficient at generating mutants with a phenotype of interest the subsequent identification of causal mutations is often cumbersome. This is particularly the case for traditional chemical mutagens such as N-ethyl-N-nitrosourea (ENU) and ethyl-methanesulfonate, which generate genome-wide point mutations and in so doing significant background noise from which a candidate gene carrying a mutation must be identified and then validated (1,2). Similarly, while ionizing radiation is a powerful tool for mutagenesis generating sufficiently small chromosomal rearrangements so that a candidate gene can be identified using approaches such as comparative genomic hybridization requires high doses of radiation to be used which generates a significant number of rearrangements, the majority of which represent background (3). Lower doses produce rearrangements of large chromosomal regions, in some cases containing hundreds of genes, complicating follow-up analysis (3). Even viruses, which can be introduced as single copy integrants into the genome, may function many megabases distal to their integration site (4). Viruses also exhibit strong insertional biases preferring to insert into active promoters and into open chromatin, which effectively reduces the compendium of genes that they can mutate (5).
Transposons are mobile genetic elements that constitute a major part of the repetitive sequence of eukaryotic genomes (6). Transposons may be classified into two groups; DNA transposons and retrotransposons. DNA transposons consist of two terminal repeats flanking a transposable element, which allows them to be mobilized and relocated to other locations in the genome by a ‘cut-and-paste’ mechanism. Retrotransposons function by a ‘copy-and-paste’ mechanism (7). In lower organisms (worms, bacteria, Drosophila), DNA transposons have been used extensively for genetic manipulation and for mutagenesis (8–10). In recent years, DNA transposons have also been used for insertional mutagenesis screens in vertebrates. In zebrafish Tol2 has gained popularity, but has limited activity in mammalian cells and in vivo (11). Other transposons including Minos have also been trialled in mammalian cells but with mixed success (12). In contrast the Tc1-family transposon Sleeping Beauty (SB) has been shown to be effective for cancer gene discovery screens in vivo (13,14), and is an active mutagen in the germline (15–17). While SB is active in some cell lines such as 293T and HeLa cells, is appears to be weakly active in embryonic stem (ES) cells (18). SB also exhibits significant ‘local hopping’, a phenomenon where a mobilized transposon re-integrates close to the donor locus (15,17,18), and to have a limited cargo capacity with mobilization being significantly reduced when elements of >3 kb are cloned between the inverted repeats/direct repeats (IR/DR) of the transposon (19). These factors, coupled with overexpression inhibition where optimal transposition is obtainable within a narrow window of transposase expression, and is repressed at high levels of expression, limits the utility of SB as a universal mutagen. Despite these factors SB-mediated screens have been successfully performed in cell culture systems (20). More recently considerable effort has been invested in developing the transposon PiggyBac (PB) from the moth Trichoplusia ni, as a tool for insertional mutagenesis (21–26). Mobilization of PB from donor loci results in a more random distribution of transposon insertion events around the genome than is obtainable with SB or Tol2. PB can mobilize large cargo containing transposons (as big as several hundred kilobases) and unlike SB expression of the PB transposase at high levels does not appear to result in overexpression inhibition (24). Another advantage of PB is that the PB transposase is still active when fused with other proteins such as the oestrogen receptor ligand-binding domain (ERT2) opening up a range of possibilities for temporally controlled mutagenesis (25). In addition to these factors PB appears to be highly active in mammalian cells generating multiple independent insertions in cells into which the transposon is introduced. Collectively these factors suggest that PB is a powerful mutagen complementary to other tools. In particular PB may be used in combination with loss of the Bloom syndrome helicase for recessive screens (23,27).
The most common method for introducing PB, or indeed other transposons, into mammalian cells is to co-transfect a plasmids expressing the transposase (‘helper’ plasmid) and another plasmid carrying a transposon (‘donor’ plasmid) (28). Once transfected into the host cells the transposase enzyme mobilizes the transposon from the donor plasmid and integrates it into the host genome. The number of transposon integration events can be controlled to some extent by titering the amount of plasmid and the ratio of the helper and donor plasmids but integration patterns are frequently complex (23). Similarly, once integrated continued expression of the transposase can re-mobilize transposon integration events generating a complex pattern of integrations in subsequent cell divisions. To overcome these limitations we have developed a novel PB system for self-inactivating insertional mutagenesis, which we have named Slingshot. The name Slingshot refers to a single shot handheld weapon. Here we show that this system can be used for ligand inducible insertional mutagenesis using 4-Hydroxytamoxifen (4-OHT) generating single stable integration events distributed genome-wide. Transposition and trapping efficiency is extremely high such that as many as 30 000 clonal insertion events can be generated from a single 10-cm plate of cells making it feasible to screen all permissive genes and regions of the genome in just a few plates of cells in culture.
MATERIALS AND METHODS
Plasmids construction
The PB Slingshot transposon with 5′ and 3′ PB terminal repeats flanking a splice acceptor-IRES, promoterless β-geo, and a cytomegalovirus early enhancer/chicken β-actin (CAG) (CAGGS) promoter was constructed using elements derived from the plasmids 5′-PTK-3′, pGTo1xr and pcDNA3.1 (25). The PB transposase estrogen receptor fusion (mPB-L3-ERT2) cDNA was obtained from Cadinanos and Bradley (25). The final Slingshot plasmid (Figure 1) was constructed with a pBluescript II SK(+) vector backbone using standard approaches and was sequenced in full. The vector has accession number GU937109.
Figure 1.
Schematic diagram of the Slingshot plasmid and the mutagenesis senarios possible with this system. The Slingshot system was constructed using a pBluescript II SK(+) vector as backbone (A). The construct can be linearized using the restriction enzyme PacI and stably integrated into a host cell genome using blastocidin selection. (B) The Slingshot transposon can be mobilized from the stably integrated Slingshot plasmid by treating cells with 4-OHT which translocates the PB-ERT2 fusion protein to the nucleus and mobilizes the transposon. (C) If the Slingshot transposon re-integrates downstream of an endogenous promoter ‘X’ it can hijack the promoter resulting in G418 resistance. (D) If the Slingshot transposon re-integrates upstream of a gene it can overexpress the gene by expression from the CAGGS promoter. (E) It is also possible for the Slingshot transposon to generate neomorphic alleles such as dominant negative alleles and loss-of-function events by generating intergenic insertions. (F) Mobilization of the transposon translocates the CAGGS promoter away from the PB-ERT2 preventing further re-mobilization events.
Cell culture and transfection
To generate stable integrants carrying the Slingshot plasmid ES cell were electroporated with 40 μg of PacI linearized Slingshot plasmid DNA; 107 E14Tg2a ES cells at 230 V, 500 mF using a BIO-RAD Gene Pulser II Electroporator. Cells were cultured on a 10-cm culture dish after transfection and selected with 15 μg/ml blasticidin (BSD) for 10 days. Individual clones were picked into 96-well plates for downstream analysis. All other cell lines were cultured in DMEM.
Trapping efficiency test
The trapping efficiency of slingshot integrated ES cells was evaluated by plating cell lines in duplicate into adjacent wells of a six-well plate and treating one well with 1 μM 4-OHT for 2 days, and the other well with 95% ethanol vehicle as a control. Cells were then cultured under G418 selection (175 μg/ml) for 8 days to select for gene trap events. Colonies were counted from 4-OHT treated and control wells to determine the transposon trapping efficiency and the level of background.
Excision PCR on DNA for 4-OHT treated cells
Excision PCR was performed using a forward and reverse PCR primer flanking the transposon in the Slingshot cassette, generating a PCR band of 526 bp if the transposon has been mobilized from the donor locus. The excision PCR was performed at 95°C 15 min/ 95°C 0.5 min, 62°C 0.5 min, 72°C 1.5 min: 40 cycles/ 72°C 5 min using following primers: FWD: 5′-AAGTGTAGCGGTCACGCTGC-3′. REV: 5′-CTCGATCACGTTCTGCTCGT-3′.
Drug resistance screen using puromycin and vincristine
To identify genes responsible for puromycin and vincristine resistance, clone PB/PB-1, which carries a single Slingshot integration, was plated onto 10 cm dishes at 106 cells per plate. Cells were treat with 1 μM 4-OHT for 2 days before selection in puromycin or vincristine media commenced. Controls were treated with 95% ethanol as a vehicle control. Selection was performed with 1 μg/ml puromycin or 10 pg/ml vincristine for 10 days. Colonies from 4-OHT treated plates were picked and genomic DNA was extracted to perform splinkerette PCR for insertion sites isolation.
Splinkerette-PCR and insertion sites analysis
For splinkerette PCR 4 μg of genomic DNA was digested overnight with 20 units of Sau3AI in a 50-μl reaction volume. After heat inactivation at 65°C for 20 min 2.5 μl of the digestion mix was ligated with annealed linker oligos overnight. This linker ligation served as template for two rounds of PCR to amplify the transposon/genome junction. Splinkerette PCR products were separated by agarose gel electrophoresis, excised from the gel and sequenced. Primer sequences for splinkerette PCR and sequencing are provided in Supplementary Table S1. Sequencing reads were analyzed using iMapper, an online web tool for insertion site analysis (29).
Western blotting
Cells were lysed in NP-40 lysis buffer with 1 mM PMSF. The protein concentration of each lysate was determined using the BCA assay (Pierce). Approximately 50 μg of each lysate was fractionated on an SDS–PAGE gel. The protein was transferred to Immobilon-P PVDF membrane (Millipore). The membrane was blocked with 5% milk in TBST for 1 h at room temperature and then incubated with primary antibodies [anti-Estrogen receptor (ER) (Santa Cruz Technology) and β-Actin (Cell signaling)] and then HRP-conjugated secondary antibodies. The protein bands were visualized using the ECL Western blotting system (GE Healthcare).
RESULTS AND DISCUSSION
Generation of the Slingshot PB system and Slingshot ES cell lines
We constructed the Slingshot PB system as shown in Figure 1. The Slingshot transposon contains a CAGGS promoter for gain-of-function mutagenesis and elements for gene trapping to generate loss-of-function mutations upon integration into the genome. The transposase, driven by the CAGGS promoter is tamoxifen inducible and has been described previously (25). After stable integration of the Slingshot cassette into the genome using blastocidin selection transposition can be initiated by the administration of 4-OHT. Mobilization, however, translocates the CAGGS promoter away from the mPB-L3-ERT2 cDNA, which should eliminate further transposase expression preventing remobilization. We electroporated 40 μg of linearized Slingshot plasmid into mouse ES cells (E14Tg2a), selected in blastocidin, and picked 18 stable integrants for further analysis.
Evaluate the trapping efficiency for the Slingshot transposon
Eighteen ES cell clones carrying the Slingshot plasmid were plated into six-well plates in duplicate. For each clone one well was treated with 4-OHT and the other well was treated with 95% ethanol as a control. As shown in Figure 2A, 10 of 18 clones showed trapping activity after 4-OHT treatment. Some clones that were found to be blastocidin resistant failed to show trapping activity, which might suggest that the Slingshot PB plasmid had been damaged during integration, or that the plasmid had integrated into a site in the genome where transposon mobilization was not permissive. Several clones (clones 3, 12, 18) show colony formation in 4-OHT treated wells but also in control wells suggesting that the Slingshot plasmid had integrated into a site where β-geo was promiscuously expressed resulting in G418 resistance. We selected clone 1 (PB/PB-1) for further analysis because it showed low background and high inducibility, which was maintained over more than 12 passages (Figure 2B). The Slingshot plasmid integration sites for this clone were characterized by Southern blotting, splinkerette PCR and only one donor site was identified and mapped on chromosome 18. The excision of the Slingshot transposon from the PB/PB-1 donor locus was verified by excision PCR in eight independent daughter clones after 4-OHT treatment and G418 selection (Figure 2C). To assess re-integration events from the PB/PB-1 Slingshot donor locus we treated PB/PB-1 cells with 4-OHT, picked 48 G418 resistance clones, and determined the position of transposon re-integration events by splinkerette PCR. From the Karyoview generated by iMapper (29) there did not appear to be local hopping from the Slingshot donor locus on chromosome 18, and insertions appeared to be widely distributed throughout the genome (Figure 2D).
Figure 2.
Slingshot is a potent insertional mutagen. The Slingshot plasmid was introduced into E14Tg2A ES cells and 18 clones were picked. (A). Mobilization of the Slingshot transposon from the Slingshot plasmid in these lines revealed the background and inducibility of each cell line for gene trap mutagenesis. (B) Clone one (PB/PB-1) showed high transposition activity and minimal background in gene trap experiments. This activity was stable of more than 12 passages. (C) Excision PCR on genomic DNA from 8 PB/PB-1 G418 resistant colonies after 4-OHT treatment. Mobilization of the transposon results in a band of 526 bp. The last lane is the PB/PB-1 control DNA without 4-OHT treatment. (D) KaryoView picture generated by iMapper of the insertion sites isolated from 48 PB/PB-1 G418 resistant clones. The red arrow indicates the original Slingshot transposon integration site.
To further characterize the transposition efficiency of the Slingshot donor in PB/PB-1 cells we performed a series of mobilization tests to calculate the trapping efficiency following 4-OHT treatment for different time periods (Supplementary Table S2). From these tests we obtained trapping efficiencies ranging from 0.2 to 0.4%, with longer 4-OHT treatment resulted in slightly higher jumping efficiency. These experiments suggested that from a confluent 10 cm dish seeded with 107 ES cell there would be sufficient insertion events to generate over 20 000 trapping events covering most permissive genes in the genome. Although the Slingshot plasmid uses the mPB-L3-ERT2 transposase which has lower activity compared with the original PB transposase, Slingshot transposition is considerably higher when compared with other studies using plasmid based delivery methods (21,24,25). It is worth noting in these experiments that we are equating the trapping efficiency of Slingshot with the mobilization efficiency. It is likely that the mobilization and re-integration efficiency of Slingshot is higher than the systems’ trapping efficiency reported in these experiments.
Drug resistance screens using the Slingshot system
The transposon used in the Slingshot system contains a CAGGS promoter and splice acceptor—polyA ‘trapping’ elements to generate both gain and loss-of-function transposition events. The trapping experiments described earlier essentially validate the loss-of-function elements of the transposon. Here we set out to test the gain-of-function capabilities of Slingshot by performing drug resistances screen in vitro using two agents, the aminonucleoside antibiotic puromycin and vincristine, an antimicrotubule spindle poison.
For the puromycin screen two experiments were performed in 10 cm plates using 8 and 20 plates seeded with 106 PB/PB-1 cells, respectively. From these experiments 44 colonies (18 and 26, respectively) were derived after puromycin selection (1 μg/ml) (Figure 3A and B). Insertion sites were isolated from these clones (Figure 3C) by splinkerette PCR and mapped to the genome with iMapper. Remarkably the insertion sites were predominantly mapped to two genomic loci. There were 21 independent insertions that mapped to chromosome 5 between 8.5 and 8.8 Mb, where the ABC Transporter genes Abcb1a and Abcb1b are localized (Figure 3D). Sixteen insertions were located upstream of Abcb1a. Five insertions were located upstream of Abcb1b before the first exon. Importantly the transposon was inserted in a sense orientation in this locus such that the CAGGS promoter will drive overexpression of these genes. Since the Abcb1a and Abcb1b genes are paralogous, having evolved as a result of gene duplication, their drug transporter activity is essentially identical. The other locus having multiple insertion sites was located on chromosome 6 between 58.54 and 58.59 Mb (Figure 3E), where five transposon insertion sites were located. These transposition events are predicted to drive overexpression of Abcg2. Abcb1a/b and Abcg2 are ABC drug transporters and here we show that puromycin is a substrate of both of these drug pumps (30).
Figure 3.
Puromycin drug resistance screen in the PB/PB-1 Slingshot ES cell line. (A) Overview of the screen. (B) PB/PB-1 ES cells were plated at a density of 106 cells per plate. Two experiments were performed using 8 and 20 plates yielding 18 and 26 puromycin resistant colonies, respectively. (C) Splinkerette PCR gel picture using 5 prime PCR primers as shown in Supplementary Table S2. Clones 1–8 are puromycin resistant clones isolated from the screen. 1′ and 2′ are control DNA from PB/PB-1. The blue arrow indicates a background PCR band (D) Twenty one independent insertion sites (indicated by blue arrowheads) were mapped to the Abcb1a/b locus on chromosome 5 between 8.5 and 8.8 Mb. (E) Five independent insertion sites (indicated in blue arrowheads) were mapped to the Abcg2 locus on chromosome 6 between 58.54 and 58.59 Mb.
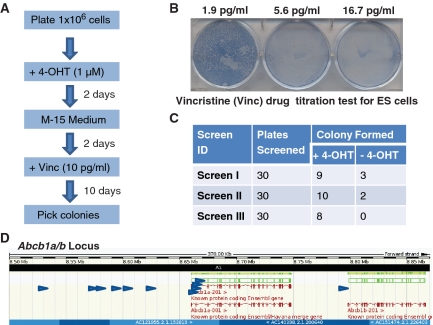
For the screen using vincristine a titration test was performed to establish the optimal concentration of drug for cell killing (Figure 4). From this test we determined that drug concentrations between 5.6 and 16.7 pg/ml efficiently killed an entire confluent well of a six-well plate within 4 days. A dose of 10 pg/ml of vincristine was selected for the screening experiments. Three screens were performed using 30 plates each time and, in total, 27 colonies were obtained (Figure 4). The insertion sites were cloned by splinkerette PCR and 11 independent insertions were mapped to the Abcb1a/b locus on the chromosome 5 between 8.5 and 8.8 Mb (Figure 4D). Therefore this screen has shown that vincristine is a substrate for the drug transporters Abcb1a/b. In both the puromycin screen and also in the vincristine screen insertions from several clones could not be mapped. This was generally due to the short sequence length of the splinkerette product or repetitive sequence that could not be mapped to a single definitive location on the genome.
Figure 4.
Vincristine (Vinc) drug resistance screen in the PB/PB-1 Slingshot ES cell line. (A) Overview of the screen. (B) Titration test for vincristine in ES cells. (C) Outcome of the drug resistance screen in PB/PB-1 cells using vincristine. (D) Eleven independent insertion sites (indicated in blue arrowheads) were mapped to the Abcb1a/b locus on chromosome 5 between 8.5 and 8.8 Mb.
Self-inactivation of the Slingshot transposon after transposition
Transposon remobilization caused by constitutive transposase expression has the potential to be a major problem in insertional mutagenesis studies. During transposition the Slingshot transposon is excised from a donor locus and re-integrates elsewhere in the genome (Figure 1F). In theory this translocates the CAGGS promoter away from the transposase shutting down further expression preventing re-mobilization of the transposon. To prove this theory we used puromycin resistant colonies generated using PB/PB-1 that had integrated upstream of Abcb1a but were G418 sensitive (Puro-1-8-14). We treated these cells with 4-OHT and then selected them in G418 to identify re-mobilization events, however none were detected illustrating that transposase activity is completely shutdown following mobilization of the Slingshot transposon (Figure 5A). The presence of transposon integration events upstream of Abcb1a was re-confirmed by genomic PCR (data not shown). Finally to prove that re-mobilization from the Abcb1a locus is possible we transfected Puro-1, -8, -14 with the mPB-L3-ERT2 plasmid by electroporation, treated cells with 1 µM 4-OHT for 2 days, and then selected them in G418 (175 µg/ml). All three cell lines treated in this way generated hundreds of colonies per plate as expected. (Figure 5A). Eight colonies were picked from mPB-L3-ERT2 transfected 4-OHT treated Puro-1, -8, -14 cultures and analyzed by PCR to confirm mobilization of the Slingshot transposon from the Abcb1a locus (data not shown). While PB-ERT2 expression was easily detectable in the PB/PB-1 clone using an anti-ER antibody we were unable to detected PB-ERT2 transposase expression in whole cell lysates from Puro-1, -8 and -14 clones further confirming that transposition from the Slingshot donor shuts down further transposase expression (Figure 5B).
Figure 5.
Slingshot is a self-inactivating transposon system. (A) Colony forming assay to test remobilization in three puromycin resistant clones, Puro-1, -8, -14. These three clones are derived from PB/PB-1 and all contain a PB transposon copy inserted upstream of the ABC transporter gene Abcb1a but are G418 sensitive. While there was no transposition after 4-OHT treatment of Puro-1, -8, -14 (top plates) transfection of a PB-ERT2 plasmid into these cell lines followed by 4-OHT treatment reveals that the Slingshot transposon can be remobilized from the Abcb1a locus (bottom plates) (B) Western Blot for PB-ERT2 transposase expression using an anti-ER antibody. Expression of PB-ERT2 is readily detectable in the PB/PB-1 control cells but completely absent from Puro-1, -8, -14.
The Slingshot transposon system is also active in somatic cell lines
To expand the utility of the Slingshot system to somatic cells, in which many cell culture screening systems have been established, we tested Slingshot in three mammalian cells lines; the human embryonic kidney cell line 293T and the Ovarian Carcinoma cell lines OVCAR-3 and PE01. All three cell lines were transfected with 40 μg of linearized Slingshot plasmid which was introduced into 107 cells by eletroporation (300 V, 800 μF) and stable integrants were selected with blastocidin (8 μg/ml for 293T, 4 μg/ml for OVCAR-3 and 3 μg/ml for PE01). Round monolayer colonies formed after 2–4 weeks of selection. Cells were treated for 2 days with 4-OHT and G418 selection was carried out for 2–3 weeks at 500 μg/ml (293T), 400 μg/ml (OVCAR-3) and 200 μg/ml (PE01) to generate data on the trapping efficiency in these cells lines. These human somatic cell lines showed considerable trapping activity after 4-OHT treatment (Figure 6). The mobilization of the Slingshot transposon from the Slingshot plasmid was confirmed by excision PCR.
Figure 6.
Slingshot is functional in a range of human somatic cell lines. Trapping efficiency assays and excision PCRs were performed in HEK293, OVCAR3 and PE01 cells stably transfected with the Slingshot plasmid. The Slingshot transposon could be mobilized in HEK293 (A–C), OVCAR3 (D–F) and also in PE01 (G–I) cells following 4-OHT administration resulting in gene trap events following analysis of 19, 18 and 11 clones from these cell lines, respectively. Excision PCR was performed in clones from each cell line pre- and post- 4-OHT treatments. N.C., negative control; P.C., positive control (DNA from the Puro-1 cell line or a mobilized human control). Human GAPDH was used as a gDNA loading control. (J) Western blot analysis of expression of the fusion protein PB-ERT2 in a highly active PB-carrying OVCAR3 clone following serial passage. The fusion protein was detected using an antibody recognizing the ERT2 domain. Parental OVCAR3 cells were used as a negative control. β-Actin was used as a loading control.
CONCLUSIONS
The recent discovery of PB, a transposon derived from the cabbage looper moth Trichoplusia ni, which is active in mammalian cells has opened up new opportunities for insertional mutagenesis (26). Compared to other insertional mutagens PB has been shown to exhibit much higher rates of transposition, less local hopping and can carry large cargo sequences. In addition the PB transposase can be fused to other sequences which has made it possible to control transposition temporally (21,24,26,28). Here we have developed a system called Slingshot, which can be used for self-inactivating insertional mutagenesis in mouse ES cells and also in a range of human somatic cell lines. We show that Slingshot can be used for genome-wide mutagenesis by performing two screens to identify mediators of resistance to the compounds puromycin and vincristine. Slingshot has several potential applications. First, as we have shown here, Slingshot represents a useful tool for genome-wide screens, secondly since the transposon is mobilized from a stable chromosomal donor by adding the ligand tamoxifen to the medium Slingshot can be used in heterogeneous populations of cells, in three dimensional culture systems, or where it is impractical to transiently transfect donor and helper plasmids into cells, and in cell lines that are hard to transfect. Thirdly, since the transposon is mobilized once and only once following the administration of tamoxifen the Slingshot system can be used to ‘barcode’ populations of cells making it possible to track the dynamics of growth of the population over time. When combined with high-throughput sequencing to read the insertion sites from a population of cells, like a molecular barcode, it should be possible to use Slingshot in synthetic genetic screens, such as those performed in yeast, opening up new opportunities for cell fate studies. We are in the process of further developing the Slingshot system so that it can be targeted into the Rosa locus, and also introduced into cells in culture in a lentivirus to facilitate the generation of single copy integrants.
ACCESSION NUMBER
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Cancer Research-UK and the Wellcome Trust (to D.J.A.). Funding for open access charge: Cancer Research UK and the Wellcome Trust.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Chen Y, Yee D, Dains K, Chatterjee A, Cavalcoli J, Schneider E, Om J, Woychik RP, Magnuson T. Genotype-based screen for ENU-induced mutations in mouse embryonic stem cells. Nat. Genet. 2000;24:314–317. doi: 10.1038/73557. [DOI] [PubMed] [Google Scholar]
- 2.Munroe RJ, Bergstrom RA, Zheng QY, Libby B, Smith R, John SW, Schimenti KJ, Browning VL, Schimenti JC. Mouse mutants from chemically mutagenized embryonic stem cells. Nat. Genet. 2000;24:318–321. doi: 10.1038/73563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schimenti JC, Libby BJ, Bergstrom RA, Wilson LA, Naf D, Tarantino LM, Alavizadeh A, Lengeling A, Bucan M. Interdigitated deletion complexes on mouse chromosome 5 induced by irradiation of embryonic stem cells. Genome Res. 2000;10:1043–1050. doi: 10.1101/gr.10.7.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Uren AG, Kool J, Berns A, van Lohuizen M. Retroviral insertional mutagenesis: past, present and future. Oncogene. 2005;24:7656–7672. doi: 10.1038/sj.onc.1209043. [DOI] [PubMed] [Google Scholar]
- 5.Mikkers H, Berns A. Retroviral insertional mutagenesis: tagging cancer pathways. Adv. Cancer Res. 2003;88:53–99. doi: 10.1016/s0065-230x(03)88304-5. [DOI] [PubMed] [Google Scholar]
- 6.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 7.Zamudio N, Bourc'his D. Transposable elements in the mammalian germline: a comfortable niche or a deadly trap? Heredity. 2010;105:92–104. doi: 10.1038/hdy.2010.53. [DOI] [PubMed] [Google Scholar]
- 8.Plasterk RH. The Tc1/mariner transposon family. Current Topics Microbiol. Immunol. 1996;204:125–143. doi: 10.1007/978-3-642-79795-8_6. [DOI] [PubMed] [Google Scholar]
- 9.Spradling AC, Stern DM, Kiss I, Roote J, Laverty T, Rubin GM. Gene disruptions using P transposable elements: an integral component of the Drosophila genome project. Proc. Natl Acad. Sci. USA. 1995;92:10824–10830. doi: 10.1073/pnas.92.24.10824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Osborne BI, Baker B. Movers and shakers: maize transposons as tools for analyzing other plant genomes. Curr. Opin. Cell Biol. 1995;7:406–413. doi: 10.1016/0955-0674(95)80097-2. [DOI] [PubMed] [Google Scholar]
- 11.Kawakami K. Tol2: a versatile gene transfer vector in vertebrates. Genome Biol. 2007;8(Suppl. 1):S7. doi: 10.1186/gb-2007-8-s1-s7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pavlopoulos A, Oehler S, Kapetanaki MG, Savakis C. The DNA transposon Minos as a tool for transgenesis and functional genomic analysis in vertebrates and invertebrates. Genome Biol. 2007;8(Suppl)(.)( 1):S2. doi: 10.1186/gb-2007-8-s1-s2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collier LS, Carlson CM, Ravimohan S, Dupuy AJ, Largaespada DA. Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature. 2005;436:272–276. doi: 10.1038/nature03681. [DOI] [PubMed] [Google Scholar]
- 14.Dupuy AJ, Akagi K, Largaespada DA, Copeland NG, Jenkins NA. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature. 2005;436:221–226. doi: 10.1038/nature03691. [DOI] [PubMed] [Google Scholar]
- 15.Fischer SE, Wienholds E, Plasterk RH. Regulated transposition of a fish transposon in the mouse germ line. Proc. Natl Acad. Sci. USA. 2001;98:6759–6764. doi: 10.1073/pnas.121569298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dupuy AJ, Fritz S, Largaespada DA. Transposition and gene disruption in the male germline of the mouse. Genesis. 2001;30:82–88. doi: 10.1002/gene.1037. [DOI] [PubMed] [Google Scholar]
- 17.Horie K, Kuroiwa A, Ikawa M, Okabe M, Kondoh G, Matsuda Y, Takeda J. Efficient chromosomal transposition of a Tc1/mariner- like transposon Sleeping Beauty in mice. Proc. Natl Acad. Sci. USA. 2001;98:9191–9196. doi: 10.1073/pnas.161071798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo G, Ivics Z, Izsvak Z, Bradley A. Chromosomal transposition of a Tc1/mariner-like element in mouse embryonic stem cells. Proc. Natl Acad. Sci. USA. 1998;95:10769–10773. doi: 10.1073/pnas.95.18.10769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Izsvak Z, Ivics Z, Plasterk RH. Sleeping Beauty, a wide host-range transposon vector for genetic transformation in vertebrates. J. Mol. Biol. 2000;302:93–102. doi: 10.1006/jmbi.2000.4047. [DOI] [PubMed] [Google Scholar]
- 20.Dasgupta M, Agarwal MK, Varley P, Lu T, Stark GR, Kandel ES. Transposon-based mutagenesis identifies short RIP1 as an activator of NFkappaB. Cell Cycle. 2008;7:2249–2256. doi: 10.4161/cc.7.14.6310. [DOI] [PubMed] [Google Scholar]
- 21.Liang Q, Kong J, Stalker J, Bradley A. Chromosomal mobilization and reintegration of Sleeping Beauty and PiggyBac transposons. Genesis. 2009;47:404–408. doi: 10.1002/dvg.20508. [DOI] [PubMed] [Google Scholar]
- 22.Yusa K, Rad R, Takeda J, Bradley A. Generation of transgene-free induced pluripotent mouse stem cells by the piggyBac transposon. Nat. Methods. 2009;6:363–369. doi: 10.1038/nmeth.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang W, Bradley A, Huang Y. A piggyBac transposon-based genome-wide library of insertionally mutated Blm-deficient murine ES cells. Genome Res. 2009;19:667–673. doi: 10.1101/gr.085621.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang W, Lin C, Lu D, Ning Z, Cox T, Melvin D, Wang X, Bradley A, Liu P. Chromosomal transposition of PiggyBac in mouse embryonic stem cells. Proc. Natl Acad. Sci. USA. 2008;105:9290–9295. doi: 10.1073/pnas.0801017105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cadinanos J, Bradley A. Generation of an inducible and optimized piggyBac transposon system. Nucleic Acids Res. 2007;35:e87. doi: 10.1093/nar/gkm446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ding S, Wu X, Li G, Han M, Zhuang Y, Xu T. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell. 2005;122:473–483. doi: 10.1016/j.cell.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 27.Luo G, Santoro IM, McDaniel LD, Nishijima I, Mills M, Youssoufian H, Vogel H, Schultz RA, Bradley A. Cancer predisposition caused by elevated mitotic recombination in Bloom mice. Nat. Genet. 2000;26:424–429. doi: 10.1038/82548. [DOI] [PubMed] [Google Scholar]
- 28.Wu SC, Meir YJ, Coates CJ, Handler AM, Pelczar P, Moisyadi S, Kaminski JM. piggyBac is a flexible and highly active transposon as compared to sleeping beauty, Tol2, and Mos1 in mammalian cells. Proc. Natl Acad. Sci. USA. 2006;103:15008–15013. doi: 10.1073/pnas.0606979103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kong J, Zhu F, Stalker J, Adams DJ. iMapper: a web application for the automated analysis and mapping of insertional mutagenesis sequence data against Ensembl genomes. Bioinformatics. 2008;24:2923–2925. doi: 10.1093/bioinformatics/btn541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Theile D, Staffen B, Weiss J. ATP-binding cassette transporters as pitfalls in selection of transgenic cells. Anal. Biochem. 399:246–250. doi: 10.1016/j.ab.2009.12.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.