Abstract
The sidechains of histidine and aspartate residues form a hydrogen bond in the active sites of many enzymes. In serine proteases, the His⋯Asp hydrogen bond of the catalytic triad is known to contribute greatly to catalysis, perhaps via the formation of a low-barrier hydrogen bond. In bovine pancreatic ribonuclease A (RNase A), the His⋯Asp dyad is composed of His119 and Asp121. Previously, site-directed mutagenesis was used to show that His119 has a fundamental role—to act as an acid during catalysis of RNA cleavage [Thompson & Raines (1994), J. Am. Chem. Soc. 116, 5467–5468]. Here, Asp121 was replaced with an asparagine or alanine residue. The crystalline structures of the two variants were determined by X-ray diffraction analysis to a resolution of 1.6 Å with an R-factor of 0.18. Replacing Asp121 with an asparagine or alanine residue does not perturb the overall conformation of the enzyme. In the structure of D121N RNase A, Nδ rather than Oδ of Asn121 faces His119. This alignment in the crystalline state is unlikely to exist in solution because catalysis by the D121N variant is not compromised severely. The steady-state kinetic parameters for catalysis by the wild-type and variant enzymes were determined for the cleavage of uridylyl(3′→5′)adenosine and poly(cytidylic acid), and for the hydrolysis of uridine 2′,3′-cyclic phosphate. Replacing Asp121 decreases the values of kcat/Km and kcat for cleavage by 101-fold (D121N) and 102-fold (D121A). Replacing Asp121 also decreases the values of kcat/Km and kcat for hydrolysis by 100.5-fold (D121N) and 10-fold (D121A), but has no other effect on the pH–rate profiles for hydrolysis. There is no evidence for the formation of a low-barrier hydrogen bond between His119 and either an aspartate or an asparagine residue at position 121. Apparently, the major role of Asp121 is to orient the proper tautomer of His119 for catalysis. Thus, the mere presence of a His⋯Asp dyad in an enzymic active site is not a mandate for its being crucial in effecting catalysis.
Keywords: catalytic dyad, catalytic triad, low-barrier hydrogen bond, pH–rate profile, ribonuclease A, serine protease, site-directed mutagenesis, X-ray crystallography
The amino acid motifs available to enzymic catalysts are diverse. Still, many enzymes fall into classes wherein great similarities exist. One well-studied class is that of the serine proteases (1). This class of enzymes has an active-site motif known as the catalytic triad, which is composed of the residues Ser⋯His⋯Asp linked by hydrogen bonds (2, 3).
The importance ascribed to the Ser⋯His⋯Asp motif is in large part due to its presence in the active sites of nonhomologous serine proteases (4). In recent years, the triad has also been found in several lipases (5, 6), Fusarium solani cutinase (7), and the Sindbis virus core protein (8). Replacing the aspartate residue in the catalytic triad of human lipoprotein lipase with a glycine is responsible for familial Type I hyperlipoproteinemia (9).
Similar to the catalytic triad, a His⋯Asp motif is found in the active sites of serine carboxypeptidase (10), acetylcholinesterase (11), phospholipase A2 (12), haloalkane dehalogenase (13), dienelactone hydrolase (14), a variety of zinc-dependent enzymes (15), and pancreatic ribonucleases (16, 17). This His⋯Asp motif is known as the “catalytic dyad”. With the exception of the pancreatic ribonucleases, enzymes containing the His⋯Asp dyad have syn-oriented carboxylates (18), though the benefit to catalysis of a syn vs anti orientation appears to be minimal (19, 20).
In all enzymes containing a catalytic triad or dyad, an hydroxyl group functions as a nucleophile. The role of the histidine residue is to abstract a proton from the hydroxyl group and thereby to increase the nucleophilicity of the oxygen. The role of the aspartate residue has been the subject of much study and debate, as has the importance of the catalytic triad in general (21).
The catalytic triad was once thought to act in a “charge relay mechanism”. Specifically, it was postulated that a proton was removed from the serine residue by histidine while another proton was being removed from histidine by aspartate (2, 22). The unperturbed pKa of aspartic acid is too low to favor such a mechanism, and the general consensus today is that the charge relay mechanism is not operative. Instead, the aspartate residue forms a hydrogen bond that could serve both to orient the histidine sidechain and to increase its pKa, enhancing its ability to abstract a proton from the serine residue.
Site-directed mutagenesis has been used to illuminate the role and importance of the aspartate residue in the catalytic triad of two serine proteases. Replacing the aspartate residue with asparagine in trypsin (23, 24) or alanine in subtilisin (25) leads to a 104-fold reduction in catalytic activity. This decrease suggests that the aspartate residue plays a critical role in catalysis. X-ray diffraction analysis of the trypsin variant revealed, however, that the catalytic histidine residue was being stabilized as an unproductive tautomer by its interaction with asparagine (26). This result suggests that one role of the aspartate residue in a Ser⋯His⋯Asp triad is to orient the proper tautomer of histidine.
Two attempts have thus far been made to determine the role of Asp121, the aspartate in the catalytic dyad of bovine pancreatic ribonuclease A [RNase A1; E.C. 3.1.27.5; (27)]. In one study, Asp121 was replaced with asparagine in a semisynthetic enzyme (28). This semisynthetic RNase A, RNase(1 – 118)•(111 – 124), consists of a noncovalent complex between residues 1 – 118 of RNase A (obtained from proteolytic digestion of RNase A), and an overlapping synthetic peptide composed of the fourteen C-terminal residues of RNase A, except with Asp121 replaced by an asparagine residue. The semisynthetic D121N analog has 5% of the catalytic activity of the analogous wild-type semisynthetic enzyme. A difficulty with interpreting the results of this study, however, is that the three-dimensional structure of the semisynthetic D121N analog exhibits numerous changes compared to that of the analogous wild-type semisynthetic enzyme (29, 30). In another study, site-directed mutagenesis was used to replace Asp121 in RNase A itself with a glutamate residue (31). The D121E enzyme has 17% of the activity of the wild-type enzyme for the hydrolysis of 0.1 mM C>p and 16% for the hydrolysis of 0.4 mM C>p. No other characterization of this variant has been reported. In related work, replacing Asp116 of the His⋯Asp dyad of angiogenin, a homolog of RNase A, with an asparagine or alanine residue was shown to increase ribonucleolytic activity by 8- and 15-fold, respectively (32).
Here, we address the role of the aspartate residue in the catalytic dyad of RNase A. RNase A catalyzes the two-step hydrolysis of the P–O5′ bond of RNA on the 3′ side of pyrimidine residues. Figure 1 depicts the classical mechanism for these reactions (33). In the transphosphorylation step, His119 of the His⋯Asp dyad acts as an acid. The slow hydrolysis of the 2′,3′-cyclic phosphate occurs separately and resembles the reverse of transphosphorylation (34, 35). It is in this second step that the action of the catalytic dyad of RNase A most resembles that of the catalytic triad. The amino acid sequences of pancreatic ribonucleases from over forty vertebrates are known (16, 17), and are evolving rapidly (36). The catalytic dyad is conserved in each of these ribonucleases, suggesting that these residues are important. We have created RNase A variants in which Asp121 is replaced with an asparagine or alanine residue. Here, we report on the three-dimensional structures of the variants, as well as their abilities to catalyze transphosphorylation and hydrolysis.
Figure 1.
Putative mechanism for the transphosphorylation (A) and hydrolysis (B) reactions catalyzed by RNase A (33). In the hydrolysis reaction, the hydrogen bonds between Asp121⋯His119⋯H2O resemble those in the renown catalytic triad of serine proteases.
EXPERIMENTAL PROCEDURES
Materials
Escherichia coli strain BL21(DE3) (F− ompT rB-mB−)(37) was from Novagen (Madison, WI). Buffers (except Tris), cUMP, and IPTG were from Sigma Chemical (St. Louis, MO). Tris was from Fisher Scientific (Pittsburgh, PA). Poly(C) was from Midland Reagent (Midland, TX). UpA was synthesized by J. E. Thompson using methods published previously (38, 39). Growth media were from Difco (Detroit, MI) or, for 10-L growths, Marcor Development (Hackensack, NJ). DNA sequences were determined with the Sequenase Version 2.0 kit from U.S. Biochemicals (Cleveland, OH). DEAE Sephadex A-25 anion exchange resin, S-Sepharose cation exchange resin, and the mono-S FPLC cation exchange column were from Pharmacia LKB (Piscataway, NJ). Bacterial terrific broth [TB; (40)] contained (in 1 L) tryptone (12 g), yeast extract (24 g), glycerol (4 mL), KH2PO4 (2.3 g), and K2HPO4 (12.5 g).
Mutagenesis
Previously, we described the construction of a bovine pancreatic cDNA library, the cloning of the cDNA that codes for RNase A, and the efficient expression of this cDNA in Escherichia coli (41). Here, the codon for Asp121 was changed to one for an asparagine or alanine residue by oligonucleotide-mediated site-directed mutagenesis (42) using the oligonucleotides: 5′–ACTACTACACGCTAGCGTTAAAGTGGACT–3′ and 5′–ACTACTACACGCTAGCGGCAAAGTGGACT–3′, where the underlined bases differ from those in the wild-type cDNA. The GCT mismatch introduced a unique, translationally silent NheI site that was used to screen for mutated plasmids. The complete sequence of the cDNA that codes for each enzyme was determined. Plasmids that direct the expression of wild-type, D121N, and D121A RNase A were transformed into E. coli BL21(DE3).
Production of RNase A in E. coli
TB (0.20 L) containing ampicillin (200 μg/mL) was inoculated with a culture frozen in aqueous glycerol (30% w/v). After growth overnight at 37 °C, cells were collected by centrifugation and suspended in 0.20 L of fresh TB. This suspension was used to inoculate 10 L of TB containing ampicillin (200 μg/mL). When A = 4 at 600 nm, IPTG was added to a final concentration of 0.5 mM. The culture was grown for an additional 3.5 h.
Cells were collected by centrifugation, resuspended in 0.10 L of cold TE buffer, and lysed by passage through a French pressure cell twice. The lysate was centrifuged at 30000g for 30 min. The resulting pellet was washed with a cold solution (0.30 L) of deoxycholate (1 mg/mL), and centrifuged again. The pellet was suspended in 0.50 L of Tris-acetic acid buffer, pH 8.0, containing urea (9 M), acetic acid (50 mM), sarcosine (40 mM), and EDTA (1 mM). Sonication was used to aid dissolution. To reduce any disulfide bonds, dithiothreitol was added to a final concentration of 50 mM. After 1 h, the solution was passed through DEAE A-25 resin (25 g) that had been equilibrated with the above resuspension solution, and acetic acid (one tenth volume) was added to the effluent. The resulting solution was dialyzed exhaustively against 20 mM acetic acid. The dialysate was centrifuged, the supernatant was diluted to 1 L, and the pH raised to 8.1 by the addition of Tris base. Reduced glutatione and oxidized glutathione were then added to final concentrations of 2.3 mM and 0.8 mM, respectively. After 24 h the resulting solution was loaded on to an S-Sepharose cation exchange column (0.20 L). The column was washed with 1 L of 50 mM HEPES buffer, pH 7.9, and the adsorbed proteins were eluted with a linear gradient (0.50 L + 0.50 L) of NaCl (0 – 0.50 M). Fractions were pooled on the basis of A at 280 nm, and the pooled fractions were diluted with buffer to a protein concentration of <10 mg/mL. The resulting solution was loaded onto an HR10/10 mono-S cation exchange FPLC column (15 cm × 1.8 cm2) that had been equilibrated with 50 mM HEPES buffer, pH 7.9. The column was washed with 50 mM HEPES buffer, pH 7.9, and the adsorbed proteins were eluted with a linear gradient (50 mL + 50 mL) of NaCl (0 M – 0.50 M) in the same buffer. The proteins eluted between 0.09 M and 0.12 M NaCl. Fractions were again pooled on the basis of A at 280 nm, which was monitored continuously. To discourage deamination, the pH of the pooled fractions was lowered to 5.5 by the addition of 20 mM acetic acid. After exhaustive dialysis against water, the proteins were concentrated with an Amicon Centriprep-10® filter, passed through a 0.45-μm filter (Schleicher & Schuell; Keene, NH), and stored at 4 °C.
The purity of the protein preparation was assessed by SDS-PAGE and native PAGE. Based on the symmetry of A280 peaks during cation exchange chromatography and the appearance of bands after electrophoresis through polyacrylamide gels, wild-type and the D121N variant were ≥99% pure and the D121A variant was ≥95% pure. The concentration of purified enzyme was determined by assuming that A = 0.72 at 277.5 nm for a 1.0 mg/mL solution (43). A 10-L growth typically yielded several hundred milligrams of purified enzyme.
Crystallization
Salt-free aqueous solutions of the D121N and D121A variants were lyophilized. Lyophilized D121N RNase A was dissolved in unbuffered water to a concentration of 60 mg/mL. Crystallization was performed in small siliconized tubes using the batch method. A solution (25 μL) of enzyme was mixed with an equal volume of 95% (v/v) 2-methyl-2,4-pentanediol (MPD) in 0.10 M MES buffer, pH 5.2, to give a final enzyme concentration of 30 mg/mL in 47.5% (v/v) MPD and 0.05 M MES buffer, pH 5.2. The pH of the buffer was determined prior to adding the MPD. Monoclinic crystals of D121N RNase A appeared after several months of incubation at 20 °C. These crystals belong to space group P21 with a = 30.52 Å, b = 38.40 Å, c = 53.31 Å, and β = 105.7°.
Lyophilized D121A RNase A was dissolved in unbuffered water to a concentration of 60 mg/mL. Crystallization was performed using the hanging drop method. Drops consisting of enzyme solution (1.5 μL), water (1.5 μL), and reservoir solution (3.0 μL) were suspended from siliconized coverslips over 0.50 mL of 2.5 M sodium acetate buffer, pH 5.3, containing saturating NaCl. Trigonal crystals of the D121A RNase A appeared within one day of incubation at 20 °C, and grew to a size of 0.5 mm × 0.5 mm × 1.0 mm after several days. These crystals belong to space group P3221 with a = b = 64.70 Å, c = 65.03 Å, and γ = 120.
X-Ray Diffraction Data Collection
All X-ray data were collected with a Siemens HI-STAR detector mounted on a Rigaku rotating anode operating at 50 kV, 90 mA and a 300 μm focal spot. The X-ray beam was collimated by double focusing mirrors. The crystal to detector distance was 12.0 cm. Data were obtained in 512 × 512 pixel format, processed with the program XDS (44, 45) and scaled using the program XSCALIBRE (G. Wesenberg and I. Rayment, unpublished results). The data set for D121N was 88% complete in the resolution range of 30 Å – 1.6 Å with an overall merging R-factor of 1.9%. The data set was 91% complete in the resolution range of 30 Å – 1.6 Å with an overall merging R-factor of 2.9%.
Structure Refinement
The starting model for the structure of D121N RNase A consisted of residues 2 – 124 of the wild-type enzyme (pdb entry 7rsa), stripped of all solvent molecules and sidechain atoms of residue 121. The model was subjected to ten cycles of least-squares refinement with the program TNT (46), yielding an initial R-factor of 23.4%. A difference fourier map (Fo – Fc) clearly showed the position of the sidechain atoms of Asn121. Manual adjustments to the model were performed by using the program FRODO (47). After several cycles of manual adjustments and least-squares refinement, the resolution was extended to 1.6 Å and water molecules were added to the model. The peak searching algorithm in TNT was used to place ordered water molecules. Water molecules with at least 1σ of 2Fo – Fc density, 3σ Fo - Fc density and within hydrogen bonding distance of the protein or of other water molecules were retained.
The structure of the D121N variant of RNase A was solved in the monoclinic space group P21 and contains only water molecules in the active site. Here, the D121N structure will be compared to that of phosphate-free wild-type RNase A, which was also crystallized from organic solvent in the P21 space group [pdb entry 7rsa; (48)]. The D121N structure will also be compared with the semisynthetic D121N RNase A analog (sD121N), which contains a sulfate ion in the active site and was crystallized from high salt in the trigonal space group P3221. The sD121N structure was solved to a resolution of 2.0 Å [pdb entry 3srn; (30)]. In a least-squares superposition, the mainchain atoms of D121N RNase A have an average RMS deviation of 0.15 Å from those of the wild-type enzyme and 0.75 Å from the sD121N analog.
The starting model for the structure of D121A RNase A consisted of residues 1 – 124 of wild-type RNase A (pdb entry 1rph), stripped of all solvent molecules and sidechain atoms of residue 121. The model was subjected to ten cycles of least-squares refinement with the program TNT (46), yielding an initial R-factor of 22.5%. A difference fourier map (Fo – Fc) clearly showed the position of the sidechain carbon of Ala121. Manual adjustments to the model were performed by using the program FRODO (47). After several cycles of manual adjustments and least-squares refinement, the resolution was extended to 1.6 Å and water molecules were added to the model. The peak searching algorithm in TNT was used to place ordered water molecules. Water molecules with at least 1σ 2Fo – Fc density, 3σ Fo – Fc density and within hydrogen bonding distance of the protein or of other water molecules were retained.
The structure of the D121A variant of RNase A was solved in the trigonal space group P3221 and contains the first example of an acetate ion in an RNase A active site. Here, the D121A structure will be compared to that of wild-type RNase A that was also crystallized from high salt in the P3221 space group and contains a bound sulfate in the active site [pdb entry 1rph; (49)]. The D121A structure will also be compared with that of the semisynthetic D121A RNase A analog (sD121A), which contains a sulfate ion in the active site and was crystallized from high salt in the P3221 space group. The sD121A structure was solved to a resolution of 2.0 Å [pdb entry 4srn; (30)]. In a least-squares superposition, the mainchain atoms of D121A RNase A have an average RMS deviation of 0.38 Å from those of the wild-type enzyme and 0.53 Å from the sD121A analog.
Enzyme Kinetics
Steady-state kinetic parameters for the cleavage of UpA and poly(C), and the hydrolysis of cUMP were determined by spectrophotometric assays. The catalytic dyad of RNase A most resembles the well-characterized catalytic triad of serine proteases during the hydrolysis reaction (Figure 1). For this reason, the steady-state kinetic parameters for the hydrolysis reaction were evaluated as a function of pH.
Assays were performed at 25 °C in a 0.10 M solution of the appropriate buffer, adjusted in pH with aqueous NaOH or HCl. Enough NaCl was added to each reaction to increase the ionic strength to I = 0.10 M, which was calculated based on the concentrations of buffer components and substrate. The buffer used for assays of the transphosphorylation of UpA (0.10 – 1.0 mM) and poly(C) (0.050 – 1.0 mM) was MES (pH 6.0). The buffers used for assays of the hydrolysis of cUMP were formic acid (pH 3.45, 4.00), acetic acid (4.48, 5.02), MES (5.53, 6.03), Bis-Tris (6.00, 7.02), BICINE (6.48, 7.97, 8.46), MOPS (6.94, 7.46), and AMPSO (9.00).
Assays were performed in a 1.0- or 0.2-cm cuvette with a Cary Model 3 spectrophotometer equipped with a Cary temperature controller. Substrate concentrations were determined by ultraviolet absorption using ε260 = 24,600 M−1cm−1 at pH 7.0 for UpA (50) and ε268 = 6200 M−1cm−1 at pH 7.8 for poly(C) (51), or by mass for cUMP. The concentrations of substrate in the assays ranged from Km/5 to 5 × Km. BSA (0.5 mg/mL) was added to the reactions containing poly(C). The cleavage of UpA was monitored at 286 nm using Δε = −755 M−1cm−1. The cleavage of poly(C) was monitored at 238 nm. The Δε at 250 nm, as calculated from the difference in molar absorptivity of the polymeric substrate and the mononucleotide cyclic phosphate product, has been reported to be 2380 M−1cm−1 at pH 6.2 (51). The Δε at 238 nm was determined to be 2792 M−1cm−1 by observing the change in absorption of a partially cleaved substrate at 250 nm and 238 nm. The hydrolysis of cUMP was monitored at either 282 nm or 293 nm. The values for the extinction coefficients determined for the hydrolysis of cUMP are listed in Table 3.
Table 3.
pH-Dependencies of the Steady-State Kinetic Parameters for the Hydrolysis of cUMP by Wild-Type RNase A, D121N RNase A, and D121A RNase A
| pH a | Wild-Type RNase A |
D121N RNase A |
D121A RNase A |
Δε282 (M−1cm−1) |
Δε293 (M−1cm−1) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| kcat (s−1) | Km (mM) | Kp (mM) | kcat (s−1) | Km (mM) | Kp (mM) | kcat (s−1) | Km (mM) | Kp (mM) | |||
| 3.45 (F) | 0.042±0.001 | 3.79±0.2 | 2.7 | 0.029±0.001 | 5.8±0.6 | 3.4 | 958 | 44.3 | |||
| 4.00 (F) | 0.156±0.003 | 2.16±0.07 | 0.79 | 0.096±0.003 | 3.1±0.2 | 0.68 | 995 | 49.1 | |||
| 4.48 (A) | 0.43±0.01 | 1.45±0.10 | 0.26 | 0.216±0.006 | 2.3±0.2 | 0.28 | 0.031±0.001 | 1.66±0.06 | 0.17 | 971 | 47.7 |
| 5.02 (A) | 1.05±0.01 | 1.34±0.03 | 0.13 | 0.47±0.02 | 1.6±0.2 | 0.10 | 0.055±0.002 | 0.957±0.007 | 0.072 | 1000 | |
| 5.53 (M) | 1.9±0.1 | 2.1±0.2 | 0.22 | 0.68±0.02 | 1.8±0.1 | 0.17 | 0.090±0.002 | 1.17±0.04 | 0.085 | 1050 | |
| 5.58 (A) | 1.96±0.04 | 1.04±0.06 | 0.090 | 0.73±0.04 | 0.9±0.1 | 0.070 | 1050 | ||||
| 6.00 (T) | 3.7±0.2 | 2.2±0.3 | 0.160 | 1140 | 53.8 | ||||||
| 6.03 (M) | 3.7±0.2 | 2.7±0.5 | 0.16 | 1.55±0.11 | 3.2±0.4 | 0.20 | 0.187±0.006 | 1.9±0.3 | 0.071 | 1140 | 53.8 |
| 6.48 (B) | 5.5±0.2 | 4.6±0.4 | 0.23 | 1180 | 55.0 | ||||||
| 6.56 (M) | 5.9±0.2 | 5.5±0.6 | 0.26 | 2.71±0.14 | 7.9±1.2 | 0.60 | 0.324±0.004 | 5.0±0.2 | 0.145 | 1180 | 55.0 |
| 6.94 (P) | 9.2±0.4 | 21±3 | 0.84 | 2.97±0.12 | 17±2 | 1.05 | 0.43±0.02 | 15±1.5 | 0.38 | 1240 | 57.3 |
| 7.02 (T) | 8.9±0.8 | 21±5 | 0.79 | 1240 | 57.3 | ||||||
| 7.46 (P) | 9.5±0.8 | 63±11 | 2.20 | 1.46±0.08 | 26±4 | 1.6 | 0.169±0.006 | 17±2 | 0.76 | 58.7 | |
| 7.97 (B) | 7.5±0.7 | 160±20 | 7.10 | 0.73±0.04 | 50±7 | 3.3 | 59.2 | ||||
| 8.46 (B) | 6.4±0.7 | 390±50 | 12.5 | 0.42±0.02 | 110±10 | 5.2 | 57.7 | ||||
| 9.00 (S) | 0.0050 b | 19.0 | 0.0010 b | 5.0 | 57.0 | ||||||
Data were obtained at 25 °C in 0.10 M buffer containing NaCl (to I = 0.10 M). Letters in parentheses denote buffer: F = formic acid, A = acetic acid, M = MES, T = Bis-Tris, B = BICINE, P = MOPS, S = AMPSO.
Represents kcat/Km rather than kcat.
Kinetic Data Analysis
Data processing and nonlinear regression analysis were performed with the program MATHCAD (MathSoft; Cambridge, MA). Initial velocity data were weighted in proportion to the magnitude of each data point. The pH–rate profiles were fitted by using the logarithm of the kinetic constants. Reported errors were generated by the asymptotic variance – covariance matrix method. Because of strong product inhibition and a relatively small change in extinction coefficient, the initial velocities for the hydrolysis of cUMP were difficult to measure reliably. For this reason the steady-state kinetic parameters for this substrate were determined by using the integrated rate equation (52, 53) to fit the complete time-course of each hydrolysis reaction. The use of the integrated rate equation provided more accurate values for the steady-state kinetic parameters as well as values for Kp, the dissociation constant for the RNase A•3′-UMP complex. Each progress curve for the hydrolysis reaction was fitted to the integrated rate equation with the inclusion of product inhibition by an iterative procedure in which the kinetic parameters were varied by hand and the fit judged by eye. The value of kcat was allowed to vary slightly from progress curve to progress curve to allow for small errors in enzyme concentration, with the values of Km and Kp being held constant. The derivative of the functions so obtained was used to determine better fitting values for the initial velocities. These velocities were then used to obtain the kinetic parameters using the Michaelis-Menten equation. If the parameters so realized differed greatly from the initial values, the process was repeated until they differed little from the prior round. Generally, one or two rounds were sufficient.
RESULTS
Oligonucleotide-mediated site-directed mutagenesis was used to change Asp121 of the His⋯Asp catalytic dyad of RNase A to an asparagine or alanine residue. The enzyme variants were produced in E. coli, purified, and crystallized. The statistics for X-ray diffraction analysis are listed in Table 1, and the structures of the active sites are shown in Figures 2 and 3.
Table 1.
X-Ray Diffraction Analysis Statistics
| Crystal data | D121N RNase A | D121A RNase A |
|---|---|---|
| Space group | P21 | P3221 |
| Cell dimensions (Å) |
a = 30.52 (1) b = 38.40 (1) c = 53.31 (1) β = 105.7°(1) |
a = 64.70 (1) b = 64.70 (1) c = 65.03 (2) |
| Protein molecules/unit cell | 2 | 6 |
| Data collection statistics | ||
| Resolution (Å) | 1.6 | 1.6 |
| No. of measured reflections | 28480 | 56287 |
| No. of unique reflections | 15756 | 27087 |
| Completeness of data (%) | 89 | 91 |
| Rmerge (I/σ > 0.33)1 | 0.019 | 0.029 |
| Average I/σ (1.7 Å – 1.6 Å) | 4.5 | 1.9 |
| Rmerge (1.7 Å – 1.6 Å) | 0.055 | 0.200 |
| Final refinement statistics | ||
| RNase A atoms | 954 | 951 |
| Solvent atoms | 102 | 103 |
| R-factor (30.0 Å – 1.6 Å)2 | 0.177 | 0.180 |
| RMS deviation from ideal geometry | ||
| Bond distances (Å) | 0.01 | 0.01 |
| Bond angles (°) | 2.1 | 2.7 |
| Average B-factors (Å2) | ||
| Protein (mainchain) | 16.5 | 23.8 |
| Protein (all atoms) | 22.1 | 28.4 |
| Acetate | 29.7 | |
| Solvent | 39.0 | 41.4 |
, where I = observed intensity and <I> = averaged intensity obtained from multiple observations of symmetry related reflections.
, where Fo and Fc are the observed and calculated structure factors, respectively.
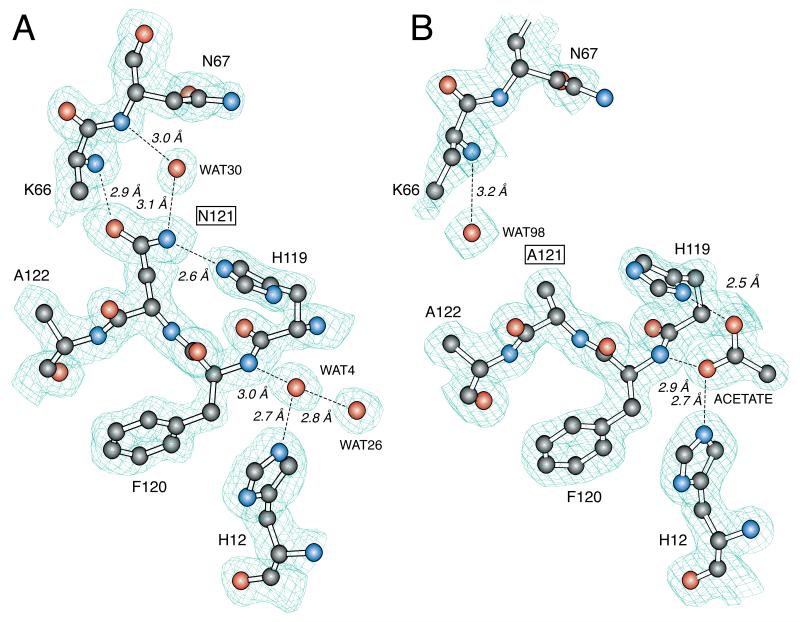
Figure 2.
Structure of the active site of crystalline D121N RNase A (A) and D121A RNase A (B). Structures were determined by X-ray diffraction analysis to a resolution of 1.6 Å. Hydrogen bonds of ≤3.2 Å are labeled. Enzymes were crystallized at pH 5.2 (D121N RNase A) or pH 5.3 (D121A RNase A).
Figure 3.
Structural differences between D121N RNase A and D121A RNase A and their semisynthetic analogs. (A) Stereoview of the overlap of the active sites of D121N RNase A (thick red lines) and the semisynthetic D121N analog [thin blue lines; (30)]. The sD121N analog contains a sulfate ion in its active site. (B) Stereoview of the overlap of the active sites of D121A RNase A (thick red lines) and the sD121A analog [thin blue lines; (30)]. D121A RNase A contains an acetate ion in its active site, and the sD121A analog contains a sulfate ion in its active site. Water molecules are depicted for the variants (red closed spheres) and semisynthetic analogs (blue open spheres). This figure was created with the program MOLSCRIPT (93).
The final model for D121N contains the complete protein (residues 1 – 124) and 102 water molecules. The R-factor for all data in the range 30 to 1.6 Å is 0.177. The RMS deviations for target geometries are 0.011 Å for bond lengths and 2.1° for bond angles. Average B-factors for the mainchain and all atoms are 16.5 and 22.1 Å2, respectively. Atomic coordinates for D121N RNase A have been deposited in the pdb with accession code 3rsd.
The final model for D121A contains the complete protein (residues 1 – 124), 100 water molecules, 2 chloride ions, and an acetate ion. The current R-factor for all data in the range 30 to 1.6 Å is 0.180. The RMS deviations for target geometries are 0.013 Å for bond lengths and 2.7° for bond angles. Average B-factors for the mainchain and all atoms are 23.8 and 28.4 Å2, respectively. Atomic coordinates for D121A RNase A have been deposited in the pdb with accession code 4rsd.
The steady-state kinetic parameters for the turnover at pH 6.0 of UpA, poly(C), and cUMP by wild-type RNase A and the D121N and D121A variants are listed in Table 2. The variations primarily affect the values of kcat. For all three substrates, the kcat for D121A RNase A is approximately 6-fold lower than that of the D121N enzyme and 20- to 50-fold lower than that of wild-type RNase A.
Table 2.
Steady-State Kinetic Parameters for Catalysis by Wild-Type RNase A, D121N RNase A, and D121A RNase A a
| Substrate | RNase A | kcat (s−1) | % | Km (mM) | % |
kcat/Km (mM−1s−1) |
% |
|---|---|---|---|---|---|---|---|
| UpA | Wild-Type | 890 ± 30 | 100 | 0.38 ± 0.03 | 100 | 2300 ± 190 | 100 |
| D121N | 190 ± 20 | 21 | 1.6 ± 0.2 | 420 | 120 ± 20 | 5 | |
| D121A | 30 ± 3 | 3.4 | 2.2 ± 0.3 | 580 | 14 ± 2 | 0.6 | |
| poly(C) | Wild-Type | 475 ± 10 | 100 | 0.14 ± 0.01 | 100 | 3300 ± 200 | 100 |
| D121N | 51 ± 2 | 11 | 0.10 ± 0.01 | 71 | 510 ± 60 | 15 | |
| D121A | 8.2 ± 0.3 | 1.7 | 0.20 ± 0.02 | 140 | 40 ± 4 | 1.2 | |
| cUMP | Wild-Type | 3.7 ± 0.2 | 100 | 2.3 ± 0.5 | 100 | 1.6 ± 0.3 | 100 |
| D121N | 1.55 ± 0.11 | 42 | 2.7 ± 0.7 | 120 | 0.58 ± 0.10 | 36 | |
| D121A | 0.187 ± 0.006 | 5.1 | 1.6 ± 0.2 | 70 | 0.11 ± 0.01 | 7 |
Data were obtained at 25 °C in 0.10 M MES buffer, pH 6.0, containing NaCl (to I = 0.10 M).
Kinetic constants for the hydrolysis of cUMP at different values of pH are listed in Table 3. These data are plotted for kcat/Km, kcat, and Kp in Figures 4 – 6. The solid lines in Figures 4 – 6 are drawn according to the following equations:
| (1) |
| (2) |
| (3) |
where [H+] is the hydrogen ion concentration; kcat/Kmint, kcatint, kcatint2, and Kpint are pH-independent constants; Ka, Kb, Kc, and Kd are the acid dissociation constants of uncomplexed enzyme; Kp is the acid dissociation constant of uncomplexed 3′-UMP; and those constants with an “s” or “p” subscript are for the enzyme•substrate or enzyme•product complex (52, 53). In eq 1, the terms associated with Kc and Kd were not included in the fit when there was no indication of their presence. As no indication for pKp,p was present, it was set at 2.0 (54). The value of pKp was set at 5.8 (55). The free enzyme values of Kp were obtained from the kcat/Km profiles. The pKa values and intrinsic parameters calculated from eq 1 – 3 are listed in Table 4.
Figure 4.

Plot of log(kcat/Km) vs pH for the hydrolysis of cUMP by wild-type RNase A (E), D121N RNase A (I), and D121A RNase A (D). The curves are nonlinear least-squares fits of the data to eq 1. To simplify comparisons, the values of kcat/Km for D121N RNase A and D121A RNase A were multiplied by scaling factors of 2.2. and 13, respectively.
Figure 6.

Plot of –log(Kp) vs pH for the hydrolysis of cUMP by wild-type RNase A (E), D121N RNase A (I), and D121A RNase A (D). The curves are nonlinear least-squares fits of the data to eq 3. No scaling factors were applied to these data.
Table 4.
Intrinsic Steady-State Kinetic Parameters and Apparent pKa’s for the Hydrolysis of cUMP by Wild-Type RNase A, D121N RNase A, and D121A RNase A a
| Parameter | Enzyme | pKa | pKb | pKc | pKd | kcat/Kmint (mM−1s−1) | |
|---|---|---|---|---|---|---|---|
| kcat/Km | Wild-Type | 5.58 ± 0.20 | 6.01 ± 0.17 | 3.80 ± 0.12 | 4.9 ± 1.8 | ||
| D121N | 5.48 ± 0.13 | 6.01 ± 0.06 | 3.80 ± 0.17 | 9.09±0.17 | 1.60 ± 0.27 | ||
| D121A | 5.68 ± 0.97 | 5.84 ± 0.96 | 0.45 ± 0.95 | ||||
|
|
|||||||
| pKa,s | pKb,s | pKc,s | kcatint1 (s−1) | kcatint2 (s−1) | |||
|
|
|||||||
| k cat | Wild-Type | 6.47 ± 0.14 | 8.63 ± 0.16 | 4.99 ± 0.15 | 10.5 ± 1.0 | 49 ± 14 | |
| D121N | 6.32 ± 0.28 | 7.44 ± 0.15 | 4.52 ± 0.29 | 3.9 ± 1.1 | 24 ± 15 | ||
| D121A | 6.66 ± 0.22 | 6.95 ± 0.21 | 4.62 ± 0.75 | 1.0 ± 0.3 | 6.5 ± 3.2 | ||
|
|
|||||||
| pKa,p | pKb,p | Kpint (mM) | |||||
|
|
|||||||
| K p | Wild-Type | 6.12 ± 0.10 | 8.53 ± 0.05 | 0.023 ± 0.002 | |||
| D121N | 6.03 ± 0.12 | 7.60 ± 0.10 | 0.020 ± 0.002 | ||||
| D121A | 6.00 ± 0.09 | 7.83 ± 0.04 | 0.013 ± 0.001 | ||||
As shown in Figure 4, wild-type RNase A, D121N RNase A, and D121A RNase A have virtually identical kcat/Km profiles except for an alkaline dip in the profile of the D121N enzyme. Both wild-type RNase A and D121N RNase A display a second low pH limb at a pH = 4.
As shown in Figure 5, wild-type RNase A, D121N RNase A, and D121A RNase A have similar kcat profiles at pH < 7, though that of the D121N enzyme is somewhat shallower than that of the others. At pH > 7, the kcat profile of the wild-type enzyme reaches a plateau, but those of the variants decrease. The kcat profiles for wild-type RNase A and D121N RNase A have slope = 1 at pH < 4.5; all three enzymes have slope < 1 between pH 4.5 and 7.0.
Figure 5.

Plot of log(kcat) vs pH for the hydrolysis of cUMP by wild-type RNase A (E), D121N RNase A (I), and D121A RNase A (D). The curves are nonlinear least-squares fits of the data to eq 2. To simplify comparisons, the values of kcat for D121N RNase A and D121A RNase A were multiplied by scaling factors of 1.6 and 16, respectively.
As shown in Figure 6, the Kp profile of D121N RNase A is similar to that of the wild-type enzyme except for a decreased Kp at high pH. The Kp of D121A RNase A is twofold less than that of the other two enzymes.
DISCUSSION
Three-Dimensional Structures
RNase A was the third enzyme (after lysozyme and carboxypeptidase) whose structure was solved by X-ray diffraction analysis (56). Despite this early precedent (57), the crystalline structures reported here are the first for active-site variants. These structures show that replacing residue Asp121 of the His⋯Asp catalytic dyad with an asparagine or alanine residue does not affect the structure of RNase A beyond residue 121. Interpretations of function based on the identity of the active-site residues alone are thus justified.
The structure of RNase A is comprised of a twisted four-stranded, antiparallel β-sheet consisting of two long central β-strands, β2 and β3 (residues 71 – 92 and 94 – 110, respectively), flanked by two shorter β-strands, β1 and β4 (residues 41 – 48 and 118 – 123, respectively). Three helicies are found in RNase A: α1 at the N-terminus (residues 3 – 13), and α2 and α3 nearly perpendicular to and at either end of the β-sheet (residues 24 – 34 and 50 – 60, respectively). Nucleotide binding occurs in a deep cleft created by α1 and β3 (58). The bases of adjacent RNA nucleotides bind in three enzymic subsites referred to as B1, B2, and B3 (59). Catalysis occurs in the P1 subsite and results in the cleavage of the P–O5′ bond specifically on the 3′-side of a pyrimidine nucleotide bound in the B1 subsite. Residues from the B1 subsite and P1 catalytic site compose the walls and floor of the groove. Gln11, Lys41, and Asn44 form one wall, and His119, Asp121, and Ala122 form the other. His12, Asn44, Thr45, and Phe120 form the floor of the B1 subsite. This discussion will focus on structural deviations observed in residues constituting the active site (Figures 2 and 3).
Gln11
The sidechain Nε2 of Gln11 can donate a hydrogen bond to a phosphoryl oxygen in the active site. Site-directed mutagenesis studies in which Gln11 is replaced by an alanine, asparagine, and histidine residue have shown that the role of Gln11 is to prevent the nonproductive binding of substrate (41). In the structure of the RNase A•uridine 2′,3′-cyclic vanadate complex, Nε2 of Gln11 forms a hydrogen bond with a nonbridging vanadyl oxygen, perhaps mimicking its interaction with the transition state (58, 60). The sidechain Nε2 of Gln11 forms a hydrogen bond with a bridging phosphoryl oxygen in an RNase A•d(CpA) complex, and with a water molecule in an RNase A•3′-CMP complex (49).
In the wild-type monoclinic structure, Nε2 of Gln11 forms a 3.0 Å hydrogen bond with a conserved water molecule in the active site, but Oε1 forms no hydrogen bonds to surrounding water molecules (48). In the structure of D121N RNase A, the sidechain of Gln11 adopts a slightly different conformation. The sidechain Nε2 of Gln11 forms a longer (3.4 Å) hydrogen bond with the conserved active-site water molecule, and Oε1 forms a 2.9 Å hydrogen bond with a new water molecule. In the sD121N analog, Nε2 of Gln11 adopts the same conformation as in wild-type RNase A, forming a 2.8 Å hydrogen bond with the conserved active-site water molecule.
In the wild-type trigonal structure, Nε2 of Gln11 forms a 2.6 Å hydrogen bond with the conserved active-site water and a 3.0 Å hydrogen bond with a sulfate oxygen bound in the active site (49). The conserved active-site water is displaced by C1 of an acetate ion in the D121A variant. The result is a small shift of Nε2 away from the acetate (due to a 30° rotation around χ3), with Oε1 forming a hydrogen bond with a new water molecule. In the sD121A structure, Nε2 of Gln11 forms a 2.8 Å hydrogen bond with the conserved active-site water molecule. But unlike in the wild-type enzyme, Nε2 in the sD121A analog does not interact with the sulfate bound in the active site.
His12
In the classical mechanism for RNase A catalysis, Nε2 of His12 acts as a base to abstract a proton from the 2′-hydroxyl group of RNA and thereby facilitate its attack on the phosphorous atom (33, 61). In all structures of RNase A in the pdb, His12 is found in the same position regardless of the crystallization conditions, solvent molecules, or ligand in the active site [(62); L. W. Schultz and R. T. Raines, unpublished results]. His12 forms a hydrogen bond between Nδ1 of its sidechain and the mainchain carbonyl of Thr45, and between Nε2 and a water molecule or a ligand in the active site. The position of His12 in the D121N or D121A variants has not changed, overlapping quite well with His12 from the sD121N and sD121A structures. In the D121N structure, Nε2 of His12 forms a 2.7 Å hydrogen bond with a water molecule in the active site. A similar 2.7 Å hydrogen bond is also found in the wild-type monoclinic structure. In the structure of the sD121N analog, Nε2 of His12 forms a 2.5 Å bond with a sulfate oxygen in the active site. In the D121A structure, Nε2 of His12 forms a 2.7 Å hydrogen bond with an acetate oxygen in the active site. This acetate oxygen is located in the same position as is the sulfate oxygen that forms a 2.8 Å hydrogen bond with Nε2 in the wild-type trigonal structure. As in the sD121N analog, His12 of the sD121A analog forms a 2.5 Å hydrogen bond with a sulfate oxygen in the active site.
Lys41
The sidechain of Lys41 has been implicated in transition state stablization, contributing 105-fold to catalysis (63). In addition, Nζ of Lys41 has been shown to form a hydrogen bond with the 2′-oxygen of 3′-CMP and to a bridging vanadyl oxygen of uridine 2′,3′-cyclic vanadate (58, 49). In the D121N and D121A variants, the sidechain of Lys41 has clearly defined density and exists in a fully extended conformation. In the wild-type monoclinic and trigonal structures, Nζ of Lys41 forms a hydrogen bond (2.6 Å and 2.7 Å, respectively) with Oδ1 of Asn44. The structures of the D121N and D121A variants have the same hydrogen bond (2.8 Å and 2.9 Å, respectively) between Nζ of Lys41 and Oδ1 of Asn44, but Nζ in the D121A enzyme makes an additional 2.5 Å hydrogen bond with a new water molecule in the active site. In the sD121N and sD121A analogs, Nζ of Lys41 also forms a hydrogen bond (2.9 Å and 3.2 Å, respectively) to Oδ1 of Asn44. As in the D121A variant, however, Nζ of Lys41 in the sD121N analog forms an additional 2.7 Å hydrogen bond with a water molecule in the active site. In the sD121A analog, Nζ also forms a 2.8 Å hydrogen bond with Oε1 of Gln11.
Residues 65 – 72
Residues 65 – 72 form a loop between α3 and β2. This loop resides near the active site and contains Lys66, which is implicated in the binding of a substrate phosphoryl group (59). Lys66 makes two interactions with Asp121 in the wild-type monoclinic RNase A structure: a 2.8 Å hydrogen bond from the mainchain N of Lys66 to Oδ2 of Asp121 and a 3.0 Å hydrogen bond from Nζ of Lys66 to the mainchain O of Asp121. In the wild-type trigonal structure, the mainchain N of Lys66 also forms a 3.3 Å hydrogen bond to Oδ2 of Asp121, but Nζ of Lys66 forms no interactions with the protein.
The hydrogen bond between the mainchain N of Lys66 and Oδ2 of Asp121 restricts conformational entropy by linking two parts of the native enzyme that are proximal in space but distal in sequence. 1H-NMR data suggest that this hydrogen bond is especially strong (64). In X-ray diffraction analyses, the length of this hydrogen bond appears to rely on the crystallization conditions, being 2.8 – 2.9 Å in salt-free crystals and 2.9 – 3.3 Å in crystals containing salt.
Movement of the loop containing residues 65 – 72 away from the active site has been proposed to be a major contributor to the reduced activity of the sD121N and sD121A analogs (30). The mainchain atoms of residues 65 – 72 in the trigonal sD121N structure have an average shift of 0.76 Å away from the wild-type mainchain. In contrast, the same atoms in the monoclinic D121N structure have an average shift of 0.45 Å away from the wild-type mainchain, suggesting that the crystallization conditions and not the amino acid substitution are responsible for the larger shift.
Other significant differences exist between the D121N and sD121N structures (Figure 3A). The hydrogen bond between the mainchain N of Lys66 and Oδ1 of Asn121 is 2.9 Å in the D121N variant, but 3.2 Å in the sD121N analog. The sidechain Nζ of Lys66 forms a 3.3 Å hydrogen bond with the mainchain O of Asn121 in the D121N variant, but a 3.5 Å hydrogen bond with Oδ1 of Asn121 in the sD121N analog. Semisynthetic F120L and F120Y analogs of RNase A in the trigonal form also form hydrogen bonds between Nζ of Lys66 and Oδ1 of Asp 121, but neither forms a hydrogen bond with the mainchain O of Asp121 (65). Another difference is more dramatic, as it involves the flipping of the Lys66 – Asn67 peptide bond. The 㗕,Ψ angles for Lys66 and Asn67 for the wild-type RNase A (−56°,−35° and −85°,1°) and the D121N variant (−55°,−39° and −82°,−5°) are similar, but those for the sD121N analog (−84°,83° and 166°,−26°) differ. In the sD121N structure, a water molecule occupies the position of the mainchain O in the D121N structure. This water molecule in the sD121N structure is only 2.1 Å (which is an unrealistic distance) from the mainchain N of Asn67. We therefore suspect that the discrepancy between the location of residues 65 – 72 in the D121N and sD121N structures is largely due to an error in electron density map interpretation.
Residues 65 – 72 in D121A RNase A are almost identical in structure to residues 65 – 72 in the sD121A analog. The loss of the hydrogen bond between the sidechain of residue 121 and the mainchain N of Lys66 has caused the mainchain atoms in residues 65 – 72 in both structures to move away from the wild-type mainchain by an average 0.7 Å. Moreover, the Lys66 sidechain forms no hydrogen bonds to the protein in either structure. The absence of this hydrogen bond is, however, not uncommon [pdb entries 1lsq and 1rnc; (66, 67)]. Although the Asp121 sidechain to Lys66 mainchain hydrogen bond appears to be important in stabilizing the structure of residues 65 – 72, it is unlikely that hydrogen bonds to the sidechain of Lys66 play a significant role in that stabilization.
His119
In the classical mechanism of RNase A catalysis (Figure 1), His119 acts as a general acid by donating a proton to the 5″-oxygen to facilitate its displacement (33, 61). The sidechain of His119 has been shown to occupy two distinct positions, denoted as A and B, which are related by a rotation around χ1 to dihedral angles of approximately 159° and −60°, respectively, and a concurrent rotation around χ2 of 180° (68, 69). The active-site histidine residue in carbonic anhydrase II undergoes a similar rotation (70). There has been considerable debate as to which position of His119 in RNase A is catalytically relevant because the imidazole nitrogens have access to the active site in both orientations (30). It has also been argued that His119 is in position A during catalysis of transphosphorylation and in position B during catalysis of hydrolysis (69). In position A, Nε2 of His119 forms a hydrogen bond with Oδ1 of Asp121, which directs Nδ1 of His119 toward the active site where it can form a hydrogen bond with a ligand (49). In the absence of a ligand, Nδ1 of His119 forms no apparent hydrogen bonds to water molecules (48). In the RNase A•3′-CMP complex, the sidechain of His119 is in position B, bringing Nε2 within 3 Å of a nonbridging phosphoryl oxygen of 3′-CMP. In position B, Nδ1 of His119 is distal from the active site. In the crystalline RNase A•d(CpA) complex, the B2 subsite is occupied by adenine and the His119 sidechain is in position A (49). If the His119 sidechain were in position B, the imidazole ring would be in direct contact with the purine base and would create a steric clash (L.W. Schultz and R.T. Raines, unpublished results). It is therefore unlikely that His119 catalyzes RNA transphosphorylation reactions from position B.
The sidechain of His119 occupies position A in the D121N structure and overlaps with His119 in the wild-type structure almost exactly, with χ1 = 160°. The sidechain Nε2 of His119 forms a 2.6 Å hydrogen bond with Nδ2 of Asn121, and Nδ1 forms no hydrogen bonds. Similarly, the sidechain of His119 occupies position A in the D121A structure but has shifted toward the active site by a 15° rotation about χ1 = 175°. In the absence of a hydrogen-bonding sidechain at residue 121, Nε2 of His119 forms no hydrogen bonds, and Nδ1 of His119 forms a 2.5 Å hydrogen bond with the acetate ion in the active site. In the sD121A (χ1 = −50°) and sD121N (χ1 = −49°) analogs, the sidechain of His119 occupies position B. Others have concluded that in the absence of a hydrogen bond with the sidechain of Asp121, His119 would prefer position B over position A (30). We too expected a priori that replacement of Asp121 would shift the B⇄A equilibrium in favor of position B because of the loss of the favorable His⋯Asp interaction. Yet in the structures of crystalline D121N RNase A and D121A RNase A, His119 is in position A (Figures 2 and 3). It appears that the position of His119 is more dependent on crystallization conditions than on the identity of residue 121. Similarly, the position of His119 has been shown to depend on the pH (65) and ionic strength (62) of the crystallization solution. Finally, unlike in serine hydrolases (71), Cε1 of His119 is not proximal to a mainchain O in either position A or position B. (Likewise, Cε1 of His12 is not near a mainchain O.)
Asn121
At a resolution of 1.6 Å, it is impossible to distinguish Nδ2 and Oδ1 in the sidechain of Asn121. It is, however, possible to distinguish these atoms based on their hydrogen bonding schemes. In our model, Oδ1 of Asn121 occupies a position similar to that of Oδ2 in wild-type RNase A, forming a 2.9 Å hydrogen bond with the mainchain N of Lys66. This orientation is sensical because it is unlikely that Nδ2 of Asn121 could be in close proximity to a mainchain nitrogen. This orientation places Nδ2 of Asn121 near Nε2 of His119. At pH 5.3 (which is the pH of the crystallization buffer), Nε2 of His119 should be protonated, thereby rendering its interaction with the Asn121 sidechain unfavorable. But, the crystals of D121N RNase A were grown in MPD, a less polar solvent than water. Accordingly, the pKa of His119 in the crystal is likely to be less than in water. We therefore propose that His119 is in the neutral imidazole state in which Nδ1 is protonated, and that Nε2 forms a 2.6 Å hydrogen bond with the sidechain of Asn121.
A water molecule forms a bridging hydrogen bond between Nδ2 of Asn121 and the mainchain N of Asn67. The Nδ2 – water hydrogen bond distance is 3.1 Å with an Nδ2 – H – O angle of 120°. This water molecule, which is conserved in several RNase A structures, could stabilize the conformation of residues 65 – 72 by linking them to residue 121. In the sD121N analog, the water molecule has moved significantly, forming a long hydrogen bond with the mainchain N of Asn67 and perhaps resulting in a shift of the residues 65 – 72 away from the active site. Still, the sidechains of Asn121 in the D121N and sD121N structures overlap almost exactly (Figure 3A).
Ala121
The mainchain near residue 121 in the D121A variant remains in the same conformation as in the trigonal wild-type structure. The sidechain Cβ of Ala121 overlaps directly with Cβ of Asp121 in the wild-type RNase structure. In the D121A variant, a water molecule takes the place of Oδ2 of Asp121 and forms a 3.2 Å hydrogen bond with the mainchain N of Lys66. This water molecule is absent from the sD121A structure. In addition, the conserved water molecule that forms a bridging hydrogen bond between Oδ1 of Asp121 and the mainchain N of Asn67 in the wild-type enzyme is missing in the D121A and sD121A structures.
Catalysis by D121N RNase A and D121A RNase A
The effect of replacing Asp121 on kcat/Km and kcat is small, but significant (Tables 2 – 4). Substitution with an asparagine residue decreases the rate constants for transphosphorylation by 101-fold and those for hydrolysis by 100.5-fold (Table 2). Substitution with an alanine residue decreases the rate constants for transphosphorylation by 102-fold and those for hydrolysis by 101-fold (Tables 2 – 4). The differential effects on transphosphorylation and hydrolysis are still greater than these values suggest because a chemical step does not limit the rate of cleavage of UpA or poly(C) (72). We conclude that the contribution of Asp121 to transphosphorylation is greater than that to hydrolysis.
The small contribution of Asp121 to catalysis belies the importance of the hydrogen bond between His119 and Asp121. This contribution is similar to that observed in an analogous study on the structure and function of phosphatidylinositol-specific phospholipase C (73). Yet, a similar His⋯Asp dyad has been proposed to form a low-barrier hydrogen bond of extraordinary strength during catalysis by the protease chymotrypsin (74, 75), though this interpretation is controversial (76). One criterion for such a bond is an 1H chemical shift of 17 – 20 ppm (77). The 1H chemical shift of Nε2H of His119 appears at a much higher field, <12 ppm (J. L. Markley, personal commun.). By this criterion, the His⋯Asp dyad of RNase A does not have a low-barrier hydrogen bond. In theory, the His⋯Asn dyad of D121N RNase A could also form a low-barrier hydrogen bond. Another criterion for such a bond is that it arise from acids with matched pKa values (79). The pKa values of acetamide [15.1; (80)] and imidazole [14.2; (81)], which are reasonable models for the sidechains of His119 and Asn121, respectively, are indeed similar. The existence of a low-barrier hydrogen bond would, however, require that the sidechain of His119 be in its neutral imidazole form. pH–rate profiles for catalysis by D121N RNase A (Figures 4 – 6) and 1H NMR titrations of His119 in the D121N variant (54) indicate that His119 titrates between its imidazole and imidazolium forms with a pKa value similar to that of His119 in wild-type RNase A. These data deny the existence of a low-barrier hydrogen bond in the His⋯Asn dyad.
The kinetics of catalysis by D121N RNase A and D121A RNase A illuminate mechanistic aspects of the B⇄A equilibrium of His119. A Coulombic interaction, like that between a cationic histidine sidechain and an anionic aspartate sidechain, has a long range. In contrast, a hydrogen bond, like that between a histidine sidechain and an aspartate or asparagine sidechain, has a short range. Dispersion interaction between a histidine sidechain and an alanine sidechain are insignificant at distance beyond van der Waals radii. We find that RNase A variants with an aspartate, asparagine, or alanine residue in position 121 have different abilities to catalyze the hydrolysis reaction (Tables 3 and 4). We therefore conclude that His119 is likely to be proximal to residue 121 during catalysis of hydrolysis, and that hydrolysis can occur with His119 in position A.
pH-Dependence of Catalysis
The overall shape of the pH–rate profiles for catalysis by RNase A can be interpreted by using a model in which the only important protonation states are those of the two active-site histidine residues, which act in concert during catalysis [Figure 1; (82)]. Accordingly, log–log plots of the pH–rate profiles for kcat/Km and kcat are expected to be bell-shaped with slopes approaching unity. The kcat/Km profiles that have been reported are indeed bell-shaped, but the shape of the kcat profiles varies with the substrate (83-86). In general, workers who have used cCMP as the substrate have found that the slope = 1 for the acidic leg of the kcat profiles; but those who have used cUMP have found that the slope < 1. The large values of Km and strong product inhibition observed for the hydrolysis step at high pH make the basic leg difficult to characterize. These factors are likely to be the cause of the discrepant values reported for kcat at high pH (83).
Our data on the pH-dependence of kcat/Km for the hydrolysis of cUMP agree with those of previous studies. [For a review, see: (87).] Our data on the pH-dependence of kcat for the hydrolysis of cUMP are most similar to those of workers who, like us, accounted for product inhibition by using the integrated rate equation (86). This approach yields a slope near one at values of pH < 4.5, as well as higher values of kcat and Km at values of pH > 7 than are reported by others (84, 85).
Values of pKa for the two histidine residues of wild-type RNase A can be derived from the kcat/Km profile (Table 4). The values agree well with results from NMR spectroscopy (88-91, 54). Differences for any given pKa value are within error, especially when recalling that when two pKa’s have similar values, the average value is much better determined from pH–rate profiles than are the individual values (92). The pKa values of the D121N and D121A enzymes that are derived from pH–rate profiles (Table 4) do not deviate significantly from those of wild-type RNase A, at least at I = 0.10 M.
Role of Asp121 in Catalysis
pH–rate profiles for catalysis by D121N RNase A are similar to those for catalysis by the wild-type enzyme (Figures 4 – 6). This similarity suggests that the same titratable groups are participating in catalysis and that these groups have similar pKa values. Yet in the structure of D121N RNase A, Nδ2 rather than Oδ1 of Asn121 is oriented toward His119. This orientation appears to stabilize a neutral form of the imidazolyl sidechain in which Nε2 is unprotonated and Nδ1 is protonated. This tautomer of His119 is likely to be ineffective, either as an acid during the transphosphorylation reaction or as a base during the hydrolysis reaction. The major role of Asp121 thus appears to be to orient the proper tautomer of His119 for catalysis. This role is similar to that assigned to the aspartate residue in the catalytic triad of trypsin (26). In the D121A enzyme, Ala121 cannot interact with the imidazolyl group of His119. Yet, D121N RNase A is a better catalyst than is D121A RNase A (Tables 2 – 4). Thus, the His119⋯Asn121 interaction observed in the crystalline D121N enzyme (Figures 2 and 3) is unlikely to exist in solution. Rather, Asn121 is likely to move such that His119 can effect catalysis.
Conclusions
Replacing Asp121, a residue that is conserved in pancreatic ribonucleases, with an asparagine or alanine residue has no effect on the overall three-dimensional structure of RNase A. Replacing Asp121 decreases the rate of transphosphorylation by ≤102-fold and the rate of hydrolysis by ≤101-fold. Thus, Asp121 of RNase A makes a significant but not a substantial contribution to catalysis. The likely role of Asp121 is simply to position the proper tautomer of His119, which Asn121 in the crystalline D121N enzyme fails to do. We speculate that the role of Asp121 in catalysis alone is unlikely to warrant its complete conservation in pancreatic ribonucleases (54).
ACKNOWLEDGMENT
We thank B. R. Kelemen, P. A. Leland, and B. M. Templer for comments on the manuscript, and Prof. J. L. Markley for advice. We are grateful to Prof. I. Rayment, Prof. H. M. Holden, and the members of their research groups for many helpful conversations and for the use of their X-ray data collection and computational facilities.
Footnotes
This work was supported by Grant GM44783 (NIH). X-ray data collection and computational facilities were supported by Grant BIR-9317398 (NSF). L.W.S. was supported by postdoctoral fellowship CA69750 (NIH). D.J.Q. was supported by Cellular and Molecular Biology Training Grant GM07215 (NIH).
- MPSO
- 3-[(1,1-dimethyl-2-hydroxyethyl)amino]-2-hydroxy-propane sulfonic acid
- BICINE
- N,N-bis(2-hydroxyethyl)glycine
- Bis-Tris
- bis(2-hydroxyethyl)iminotris(hydroxymethyl)methane
- BSA
- bovine serum albumin
- cCMP
- cytidine 2′,3′-cyclic phosphate (otherwise C>p)
- cUMP
- uridine 2′,3′-cyclic phosphate (otherwise U>p)
- DEAE
- diethylaminoethyl
- EDTA
- ethylenediaminetetraacetic acid
- HEPES
- N-(2-hydroxyethyl)piperazine-N’-(2-ethanesulfonic acid)
- IPTG
- isopropyl β-D-thiogalactopyranoside
- MES
- 2-(N-morpholino)ethanesulfonic acid
- MOPS
- 3-(N-morpholino)propanesulfonic acid
- MPD
- 2-methyl-2,4-pentanediol
- poly(C)
- poly(cytidylic acid)
- PAGE
- polyacrylamide gel electrophoresis
- pdb
- Protein Data Bank, which is maintained by the Brookhaven National Laboratory (http://pdb.pdb.bnl.gov/)
- ppm
- parts per million
- RNase A
- bovine pancreatic ribonuclease A
- sD121A RNase A
- semisynthetic RNase A consisting of a noncovalent complex between residues 1 – 118 and residues 111 – 124, with Asp121 replaced by an alanine residue
- sD121N RNase A
- semisynthetic RNase A consisting of a noncovalent complex between residues 1 – 118 and residues 111 – 124, with Asp121 replaced by an asparagine residue
- SDS
- sodium dodecyl sulfate
- TB
- terrific broth
- Tris
- tris(hydroxymethyl)methane
- 3′-UMP
- uridine 3′-phosphate (otherwise Up)
- UpA
- uridylyl(3′→5′)adenosine.
REFERENCES
- 1.Neurath H. In: Proteolytic Enzymes: A Practical Approach. Beynon RJ, Bond JS, editors. IRL Press; New York: 1989. pp. 1–13. [Google Scholar]
- 2.Blow DM, Birktoft JJ, Hartley BS. Nature. 1969;221:337–340. doi: 10.1038/221337a0. [DOI] [PubMed] [Google Scholar]
- 3.Blow DM. Trends Biochem. Sci. 1997;22:405–408. doi: 10.1016/s0968-0004(97)01115-8. [DOI] [PubMed] [Google Scholar]
- 4.Neurath H. Science. 1984;224:350–357. doi: 10.1126/science.6369538. [DOI] [PubMed] [Google Scholar]
- 5.Brady L, Brzozowski AM, Derewenda ZS, Dodson E, Dodson G, Tolley S, Turkenburg JH, Christiansen L, Huge-Jensen B, Norskov L, L. T, Menge U. Nature. 1990;343:767–770. doi: 10.1038/343767a0. [DOI] [PubMed] [Google Scholar]
- 6.Winkler FK, D’Arcy A, Hunziker W. Nature. 1990;343:771–774. doi: 10.1038/343771a0. [DOI] [PubMed] [Google Scholar]
- 7.Martinez C, De Geus P, Lauwereys M, Matthyssens G, Cambillau C. Nature. 1992;356:615–618. doi: 10.1038/356615a0. [DOI] [PubMed] [Google Scholar]
- 8.Choi H-K, Tong L, Minor W, Dumas P, Boege U, Rossmann MG, Wengler G. Nature. 1991;354:37–43. doi: 10.1038/354037a0. [DOI] [PubMed] [Google Scholar]
- 9.Faustinella F, Chang A, Van Biervliet JP, Rosseneu M, Vinaimont N, Smith LC, Chen S-H, Chan L. J. Biol. Chem. 1991;266:14418–14424. [PubMed] [Google Scholar]
- 10.Liao D-I, Breddam K, Sweet RM, Bullock T, Remington SJ. Biochemistry. 1992;31:9796–9812. doi: 10.1021/bi00155a037. [DOI] [PubMed] [Google Scholar]
- 11.Sussman JL, Harel M, Frolow F, Oefner C, Goldman A, Toker L, Silman I. Science. 1991;253:872–879. doi: 10.1126/science.1678899. [DOI] [PubMed] [Google Scholar]
- 12.Li Y, Tsai M-D. J. Am. Chem. Soc. 1993;115:8523–8526. [Google Scholar]
- 13.Verschueren KHG, Seljée F, Rozeboom HJ, Kalk KH, Dijkstra BW. Nature. 1993;363:693–698. doi: 10.1038/363693a0. [DOI] [PubMed] [Google Scholar]
- 14.Pathak D, Ollis D. J. Mol. Biol. 1990;214:497–525. doi: 10.1016/0022-2836(90)90196-s. [DOI] [PubMed] [Google Scholar]
- 15.Christianson DW, Alexander RS. J. Am. Chem. Soc. 1989;111:6412–6419. [Google Scholar]
- 16.Beintema JJ. Life Chem. Rep. 1987;4:333–389. [Google Scholar]
- 17.Beintema JJ, Schüller C, Irie M, Carsana A. Prog. Biophys. Molec. Biol. 1988;51:165–192. doi: 10.1016/0079-6107(88)90001-6. [DOI] [PubMed] [Google Scholar]
- 18.Gandour RD. Bioorg. Chem. 1981;10:169–176. [Google Scholar]
- 19.Zimmerman SC, Cramer KD. J. Am. Chem. Soc. 1988;110:5906–5908. [Google Scholar]
- 20.Cramer KD, Zimmerman SC. J. Am. Chem. Soc. 1990;112:3680–3682. [Google Scholar]
- 21.Schowen RL. In: Mechanistic Principles of Enzyme Activity. Liebman JF, Greenberg A, editors. VCH Publishers; New York: 1988. pp. 119–168. [Google Scholar]
- 22.Umeyama H, Nakagawa S, Fujii T. Chem. Pharm. Bull. 1979;27:974–980. doi: 10.1248/cpb.27.974. [DOI] [PubMed] [Google Scholar]
- 23.Craik CS, Largman C, Fletcher T, Roczniak S, Barr PJ, Fletterick R, Rutter WJ. Science. 1985;228:291–297. doi: 10.1126/science.3838593. [DOI] [PubMed] [Google Scholar]
- 24.Corey DR, Craik CS. J. Am. Chem. Soc. 1992;114:1784–1790. [Google Scholar]
- 25.Carter P, Wells JA. Nature. 1988;332:564–568. doi: 10.1038/332564a0. [DOI] [PubMed] [Google Scholar]
- 26.Sprang S, Standing T, Fletterick RJ, Stroud RM, Finer-Moore JF, Xuong NH, Hamlin R, Rutter WJ, Craik CS. Science. 1987;237:905–909. doi: 10.1126/science.3112942. [DOI] [PubMed] [Google Scholar]
- 27.Raines RT. Chem. Rev. 1998;98 In Press. [Google Scholar]
- 28.Stern MS, Doscher MS. FEBS Lett. 1984;171:253–256. doi: 10.1016/0014-5793(84)80498-6. [DOI] [PubMed] [Google Scholar]
- 29.Cederholm MT, Stuckey JA, Doscher MS, Lee L. Proc. Natl. Acad. Sci. U.S.A. 1991;88:8116–8120. doi: 10.1073/pnas.88.18.8116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Mel VSJ, Martin PD, Doscher MS, Edwards BFP. J. Biol. Chem. 1992;267:247–256. [PubMed] [Google Scholar]
- 31.Trautwein K, Holliger P, Stackhouse J, Benner SA. FEBS Lett. 1991;281:275–277. doi: 10.1016/0014-5793(91)80410-5. [DOI] [PubMed] [Google Scholar]
- 32.Harper JW, Vallee BL. Proc. Natl. Acad. Sci. U.S.A. 1988;88:7139–7143. doi: 10.1073/pnas.85.19.7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Findlay D, Herries DG, Mathias AP, Rabin BR, Ross CA. Nature. 1961;190:781–784. doi: 10.1038/190781a0. [DOI] [PubMed] [Google Scholar]
- 34.Cuchillo CM, Parés X, Guasch A, Barman T, Travers F, Nogués MV. FEBS Lett. 1993;333:207–210. doi: 10.1016/0014-5793(93)80654-d. [DOI] [PubMed] [Google Scholar]
- 35.Thompson JE, Venegas FD, Raines RT. Biochemistry. 1994;33:7408–7414. doi: 10.1021/bi00189a047. [DOI] [PubMed] [Google Scholar]
- 36.Doolittle RF. Protein Sci. 1992;1:191–200. doi: 10.1002/pro.5560010201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Studier FW, Moffatt BA. J. Mol. Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 38.Ogilvie KK, Beaucage SL, Schifman AL, Theriault NY, Sadana KL. Can. J. Chem. 1978;56:2768–2780. [Google Scholar]
- 39.Beaucage SL, Caruthers MH. Tetrahedron Lett. 1981;22:1859–1862. [Google Scholar]
- 40.Tartof KD, Hobbs CA. Bethesda Res. Lab. Focus. 1987;9:12. [Google Scholar]
- 41.delCardayré SB, Ribó M, Yokel EM, Quirk DJ, Rutter WJ, Raines RT. Protein Eng. 1995;8:261–273. doi: 10.1093/protein/8.3.261. [DOI] [PubMed] [Google Scholar]
- 42.Kunkel TA, Roberts JD, Zakour RA. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 43.Sela M, Anfinsen CB, Harrington WF. Biochim. Biophys. Acta. 1957;26:502–512. doi: 10.1016/0006-3002(57)90096-3. [DOI] [PubMed] [Google Scholar]
- 44.Kabsch W. J. Appl Crystallogr. 1988;21:67–71. [Google Scholar]
- 45.Kabsch W. J. Appl. Crystallogr. 1988;21:916–924. [Google Scholar]
- 46.Tronrud DE, Ten-Eyck LF, Matthews BW. Acta Crystallogr., Sect. A. 1987;43:489–501. [Google Scholar]
- 47.Jones TA. Methods Enzymol. 1985;115:157–171. doi: 10.1016/0076-6879(85)15014-7. [DOI] [PubMed] [Google Scholar]
- 48.Wlodawer A, Anders LA, Sjölin L, Gilliland GL. Biochemistry. 1988;27:2705–2717. doi: 10.1021/bi00408a010. [DOI] [PubMed] [Google Scholar]
- 49.Zegers I, Maes D, Thi MH, Poortmans F, Palmer RA, Wyns L. Protein Sci. 1994;3:2322–2339. doi: 10.1002/pro.5560031217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Warshaw MM, Tinoco I. J. Mol. Biol. 1966;20:29–38. doi: 10.1016/0022-2836(66)90115-x. [DOI] [PubMed] [Google Scholar]
- 51.Yakovlev GI, Moiseyev GP, Bezborodova SI, Both V, Sevcik J. Eur. J. Biochem. 1992;204:187–190. doi: 10.1111/j.1432-1033.1992.tb16622.x. [DOI] [PubMed] [Google Scholar]
- 52.Segel IH. Enzyme Kinetics. John Wiley & Sons; New York, NY: 1975. [Google Scholar]
- 53.Cornish-Bowden A. Fundamentals of Enzyme Kinetics. Portland Press; London: 1995. [Google Scholar]
- 54.Quirk DJ. Ph.D. Thesis. University of Wisconsin–Madison; 1996. [Google Scholar]
- 55.Tanokura M. J. Biochem. 1983;94:1621–1630. [PubMed] [Google Scholar]
- 56.Kartha G, Bello J, Harker D. Nature. 1967;213:862–865. doi: 10.1038/213862a0. [DOI] [PubMed] [Google Scholar]
- 57.Gilliland GL. In: Ribonucleases: Structures and Functions. D’Alessio G, Riordan JF, editors. Academic Press; New York: 1997. pp. 306–341. [Google Scholar]
- 58.Wlodawer A, Miller M, Sjölin L. Proc. Natl. Acad. Sci. U.S.A. 1983;80:3628–3631. doi: 10.1073/pnas.80.12.3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Parés X, Nogués MV, de Llorens R, Cuchillo CM. Essays Biochem. 1991;26:89–103. [PubMed] [Google Scholar]
- 60.Ladner JE, Wladkowski BD, Svensson LA, Sjölin L, Gilliland GL. Acta Crystallogr., Sect. D. 1997;53:290–301. doi: 10.1107/S090744499601582X. [DOI] [PubMed] [Google Scholar]
- 61.Thompson JE, Raines RT. J. Am. Chem. Soc. 1994;116:5467–5468. doi: 10.1021/ja00091a060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fedorov AA, Joseph-McCarthy D, Fedorov E, Sirakova D, Graf I, Almo SC. Biochemistry. 1996;35:15962–15979. doi: 10.1021/bi961533g. [DOI] [PubMed] [Google Scholar]
- 63.Messmore JM, Fuchs DN, Raines RT. J. Am. Chem. Soc. 1995;117:8057–8060. doi: 10.1021/ja00136a001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baker WR, Kintanar A. Arch. Biochem. Biophys. 1996;327:189–199. doi: 10.1006/abbi.1996.0108. [DOI] [PubMed] [Google Scholar]
- 65.de Mel VSJ, Doscher MS, Martin PD, Edwards BFP. FEBS Leters. 1994;349:155–160. doi: 10.1016/0014-5793(94)00664-4. [DOI] [PubMed] [Google Scholar]
- 66.Capasso S, Giordano F, Mattia CA, Mazzarella L, Zagari A. Biopolymers. 1983;22:327–332. doi: 10.1002/bip.360220142. [DOI] [PubMed] [Google Scholar]
- 67.Aguilar CF, Thomas PJ, Mills A, Moss DS, Palmer RA. J. Mol. Biol. 1992;224:265–267. doi: 10.1016/0022-2836(92)90589-c. [DOI] [PubMed] [Google Scholar]
- 68.Borkakoti N, Moss DS, Palmer RA. Acta Crystallogr. B. 1982;38:2210–2217. [Google Scholar]
- 69.Borkakoti N. Eur. J. Biochem. 1983;132:89–94. doi: 10.1111/j.1432-1033.1983.tb07329.x. [DOI] [PubMed] [Google Scholar]
- 70.Nair SK, Christianson DW. J. Am. Chem. Soc. 1991;113:9455–9458. [Google Scholar]
- 71.Derewenda ZS, Derewenda U, Kobos PM. J. Mol. Biol. 1994;241:83–93. doi: 10.1006/jmbi.1994.1475. [DOI] [PubMed] [Google Scholar]
- 72.Thompson JE, Kutateladze TG, Schuster MC, Venegas FD, Messmore JM, Raines RT. Bioorg. Chem. 1995;23:471–481. doi: 10.1006/bioo.1995.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gässler CS, Ryan M, Liu T, Griffith OH, Heinz DW. Biochemistry. 1997;36:12802–12813. doi: 10.1021/bi971102d. [DOI] [PubMed] [Google Scholar]
- 74.Frey PA, Whitt SA, Tobin JB. Science. 1994;264:1927–1930. doi: 10.1126/science.7661899. [DOI] [PubMed] [Google Scholar]
- 75.Cassidy CS, Lin J, Frey PA. Biochemistry. 1997;36:4576–4584. doi: 10.1021/bi962013o. [DOI] [PubMed] [Google Scholar]
- 76.Ash EL, Sudmeier JL, De Fabo EC, Bachovchin WW. Science. 1997;278:1128–1132. doi: 10.1126/science.278.5340.1128. [DOI] [PubMed] [Google Scholar]
- 77.Cleland WW, Kreevoy MM. Science. 1994;264:1887–1890. doi: 10.1126/science.8009219. [DOI] [PubMed] [Google Scholar]
- 78.Patel DJ, Woodward CK, Bovey FA. Proc. Natl. Acad. Sci. U.S.A. 1972;69:599–602. doi: 10.1073/pnas.69.3.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cleland WW. Biochemistry. 1992;31:317–319. doi: 10.1021/bi00117a001. [DOI] [PubMed] [Google Scholar]
- 80.Bordwell FG. Acc. Chem. Res. 1988;21:456–463. [Google Scholar]
- 81.Walba H, Isensee RW. J. Am. Chem. Soc. 1955;77:5488–5492. [Google Scholar]
- 82.Sowa GA, Hengge AC, Cleland WW. J. Am. Chem. Soc. 1997;119:2319–2320. [Google Scholar]
- 83.Herries DG, Mathias AP, Rabin BR. Biochem. J. 1962;85:127–134. doi: 10.1042/bj0850127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.delRosario EJ, Hammes GG. Biochemistry. 1969;8:1884–1889. doi: 10.1021/bi00833a017. [DOI] [PubMed] [Google Scholar]
- 85.Machuga E, Klapper MH. Biochim. Biophys. Acta. 1977;481:526–41. doi: 10.1016/0005-2744(77)90285-6. [DOI] [PubMed] [Google Scholar]
- 86.Eftink MR, Biltonen RL. Biochemistry. 1983;22:5123–5134. doi: 10.1021/bi00291a011. [DOI] [PubMed] [Google Scholar]
- 87.Richards FM, Wyckoff HW. The Enzymes IV. 1971:647–806. [Google Scholar]
- 88.Meadows DH, Roberts GCK, Jardetzky O. J. Mol. Biol. 1969;45:491–511. doi: 10.1016/0022-2836(69)90308-8. [DOI] [PubMed] [Google Scholar]
- 89.Roberts GCK, Dennis EA, Meadows DH, Cohen JS, Jardetzky O. Proc. Natl. Acad. Sci. U.S.A. 1969;62:1151–1158. doi: 10.1073/pnas.62.4.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Markley JL, Finkenstadt WR. Biochemistry. 1975;14:3562–3566. doi: 10.1021/bi00687a008. [DOI] [PubMed] [Google Scholar]
- 91.Patel DJ, Canuel LL, Bovey FA. Biopolymers. 1975;14:987–997. doi: 10.1002/bip.1975.360140508. [DOI] [PubMed] [Google Scholar]
- 92.Brocklehurst K. Protein Eng. 1994;7:291–299. doi: 10.1093/protein/7.3.291. [DOI] [PubMed] [Google Scholar]
- 93.Kraulis PJ. J. Appl. Crystallogr. 1991;24:946–950. [Google Scholar]