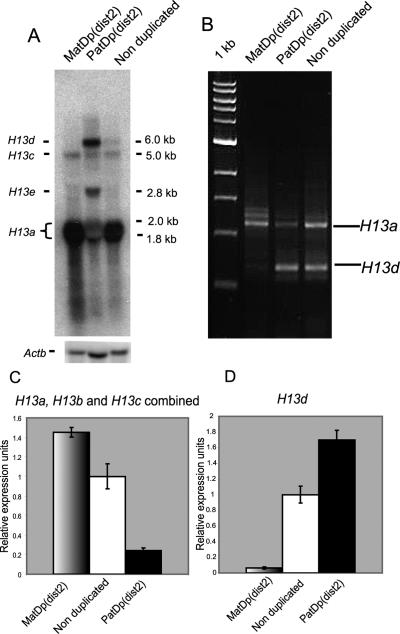
Figure 3.
Disrupted polyadenylation in mice carrying uniparental partial disomies. (A) Northern generated with total RNA extracted from neonatal brains of mice carrying uniparental partial duplications for distal Chromosome 2. Nonduplicated mice are either homozygous, heterozygous, or noncarriers of the T11H translocation but in all cases will inherit distal Chromosome 2 biparentally. The blot was hybridized with probe #3 from Figure 1A. One biological replicate was performed. Note that H13a transcripts are not completely absent following paternal inheritance, but H13d transcripts are not detected in MatDp(dist2). (B) Qualitative 3′ RACE assay, using the same gene-specific primer employed in Figure 2B. Two biological replicates gave similar results (data not shown). (C) Combined expression of transcripts extending through the DMR (H13a–c) in neonatal brains measured by QRT–PCR. Data are presented as ratios of the mean Ct value in relation to the mean Ct value for β-actin for each genotype. Microarray expression data confirmed that this endogenous control gene did not change significantly between MatDp(dist2) and PatDp(dist2) samples (Supplemental Fig. 2). Ct values are transformed as described (Pfaffl 2001) and are presented in relation to nonduplicated controls that are set to a value of 1. Amplifications were performed in quadruplicate (n = 4). Error bars represent the standard error of the mean. The amplified region spanned exons 6 and 7. (D) Expression of H13d transcripts measured by QRT–PCR. The amplified region lies within 500 bp of the H13d polyA site. Data are presented as in C.