Abstract
The importin-α/β complex and the GTPase Ran mediate nuclear import of proteins with a classical nuclear localization signal. Although Ran has been implicated also in a variety of other processes, such as cell cycle progression, a direct function of Ran has so far only been demonstrated for importin-mediated nuclear import. We have now identified an entire class of ∼20 potential Ran targets that share a sequence motif related to the Ran-binding site of importin-β. We have confirmed specific RanGTP binding for some of them, namely for two novel factors, RanBP7 and RanBP8, for CAS, Pse1p, and Msn5p, and for the cell cycle regulator Cse1p from Saccharomyces cerevisiae. We have studied RanBP7 in more detail. Similar to importin-β, it prevents the activation of Ran's GTPase by RanGAP1 and inhibits nucleotide exchange on RanGTP. RanBP7 binds directly to nuclear pore complexes where it competes for binding sites with importin-β, transportin, and apparently also with the mediators of mRNA and U snRNA export. Furthermore, we provide evidence for a Ran-dependent transport cycle of RanBP7 and demonstrate that RanBP7 can cross the nuclear envelope rapidly and in both directions. On the basis of these results, we propose that RanBP7 might represent a nuclear transport factor that carries an as yet unknown cargo, which could apply as well for this entire class of related RanGTP-binding proteins.
The nuclear pore complexes (NPC)1 are the sites where the exchange of macromolecules between nucleus and cytoplasm occurs (Feldherr et al., 1984). Transport through the NPCs is bidirectional and comprises a multitude of substrates. Small molecules can passively diffuse through the NPC. The transport of proteins and RNAs >40–60 kD is, however, generally an active process, i.e., it is energy dependent (Newmeyer et al., 1986) and mediated by saturable transport receptors (Goldfarb et al., 1986; Michaud and Goldfarb, 1991; Jarmolowski et al., 1994). To accomplish multiple rounds of transport, these transport receptors are thought to shuttle between nucleus and cytoplasm (Goldfarb et al., 1986). An import receptor, for example, has to bind its import substrate initially in the cytoplasm, release it after NPC passage into the nucleus, and return to the cytoplasm without the cargo. Conversely, an export factor has to bind the export substrate only in the nucleus and on the way out. This model predicts asymmetry in these transport cycles and implies that the binding of the transport receptor to its cargo is regulated by the different environments of nucleus and cytoplasm.
The nuclear import of proteins with a classical nuclear localization signal (NLS) is to date the best characterized nucleocytoplasmic transport pathway (for reviews see Görlich and Mattaj, 1996; Koepp and Silver, 1996; Schlenstedt, 1996). The signal contains one or more clusters of basic amino acids (for review see Dingwall and Laskey, 1991) and is recognized by the importin-α/β complex (for references see Sweet and Gerace, 1995; Panté and Aebi, 1996). The α subunit provides the NLS binding site (Imamoto et al., 1995; Weis et al., 1995) and is therefore also called the NLS receptor (Adam and Gerace, 1991). The β subunit accounts for the interaction with the NPC (Görlich et al., 1995; Moroianu et al., 1995) and carries importin-α with the NLS substrate into the nucleus. The translocation into the nucleus is terminated by the disassembly of the importin complex, and both subunits are returned probably separately to the cytoplasm. Importin-α interacts with -β via its importin-β binding domain (IBB domain; Görlich et al., 1996a ; Weis et al., 1996a ). Binding to importin-β with an IBB domain is sufficient for nuclear entry, and the IBB domain can therefore be regarded as the nuclear targeting signal of importin-α. The export domain of importin-α has not yet been identified, but it is distinct from the IBB domain.
The small GTPase Ran (Drivas et al., 1990; Bischoff and Ponstingl, 1991b ; Belhumeur et al., 1993) plays a key role in NLS-dependent protein import (Melchior et al., 1993; Moore and Blobel, 1993). GTP hydrolysis by Ran is absolutely essential for import (Melchior et al., 1993; Moore and Blobel, 1993; Schlenstedt et al., 1995a ; Palacios et al., 1996) and is possibly even its sole source of energy (Weis et al., 1996b ). Although the molecular mechanism of import is far from being understood, it appears that Ran fulfils at least two distinct functions in this process: first, Ran's GTP cycle probably drives translocation into the nucleus (Melchior et al., 1993; Moore and Blobel, 1993; Weis et al., 1996b ), which appears to involve the binding of (cytoplasmic) RanGDP to the NPC, followed by nucleotide exchange and GTP hydrolysis, but it does not involve binding of RanGTP to importin-β (Görlich et al., 1996b ). Unfortunately, nothing is known of how Ran's GTP cycle would translate into a directed movement through the NPC. Secondly, Ran regulates the interaction between importin-α and -β (Rexach and Blobel, 1995; Chi et al., 1996; Görlich et al., 1996b ). Binding of RanGTP to importin-β disassembles the importin-α/β complex at the nuclear side of the NPC, thereby terminating translocation (Görlich et al., 1996b ). The asymmetric distribution of Ran's principal GDP/GTP exchange factor (RCC1; Bischoff and Ponstingl, 1991a ) and GTPase activating protein (RanGAP1, or RNA1 in yeast; Bischoff et al., 1995a ; Becker et al., 1995) crucially determines where the importin heterodimer can form and where it is forced to dissociate. RCC1 is a nuclear, chromatin-bound protein (Ohtsubo et al., 1987, 1989) that generates RanGTP in the nucleus, whereas free RanGTP is depleted from the cytoplasm by RanGAP1, which is excluded from the nucleoplasm (Hopper et al., 1990; Matunis et al., 1996; Mahajan et. al, 1997). Thus, low RanGTP levels in the cytoplasm allow importin-α to bind -β, and the high RanGTP concentration in the nuclear compartment dissociates the importin complex. The concentration of free RanGTP can, in this model, be regarded as a marker for cytoplasmic identity (low RanGTP) and nuclear identity (high RanGTP), which is “sensed” by the Ran-binding site in importin-β.
It is likely that at least some properties of importin-β are shared by the mediators of the other nucleocytoplasmic transport pathways. This is emphasized by the recent identification of the importin-β–related transportin (Pollard et al., 1996) as an import receptor recognizing the M9 domain, the nuclear targeting signal in hnRNP A1 (Michael et al., 1995), and of yeast transportin (Kap 104p) as an import receptor for mRNA binding proteins (Aitchison et al., 1996). Furthermore, importin-β or its NPC-binding domain cross-compete with other pathways, such as M9-dependent import, NES-mediated nuclear export, and the export of mRNA and U snRNA (Kutay et al., 1997). This would suggest that these other transport receptors share at least some binding sites at the NPC and take a similar path through the nuclear pore complex as importin-β.
In addition to importin-β, a number of other Ran-binding proteins are detectable in eukaryotic cells, e.g., in overlay blots using Ran γ-[32P]GTP as a probe. These can be grouped into two classes (Lounsbury et al., 1994, 1996): first, those with a RanBP1 homology domain including the Ran binding protein 1 (RanBP1) itself (Coutavas et al., 1993; Bischoff et al., 1995b ) and the nuclear pore protein RanBP2, which has four RanBP1 homology domains (Wu et al., 1995; Yokoyama et al., 1995). Their binding to Ran can be competed by RanBP1. Second, importin-β and so far unidentified protein(s) of ∼120 kD whose Ran-binding is competed by importin-β but not by excess of RanBP1 (Lounsbury et al., 1994, 1996). Both RanBP1 and importin-β inhibit the nucleotide exchange on RanGTP (Coutavas et al., 1993; Lounsbury et al., 1994, 1996; Bischoff et al., 1995b ; Görlich et al., 1996b ). However, they do not cross-compete with each other for Ran binding but instead bind to different, nonoverlapping sites on Ran (Chi et al., 1996; Kutay et al., 1997; Lounsbury and Macara, 1997). Another striking difference is that RanBP1 facilitates the activation of Ran's GTPase by RanGAP1 (Beddow et al., 1995; Bischoff et al., 1995b ), whereas the importin-β/RanGTP complex is entirely GAP resistant (Floer and Blobel, 1996; Görlich et al., 1996b ).
Although a direct involvement of Ran has so far only been demonstrated in the importin-dependent transport pathway, perturbations in the Ran system have severe effects on a great variety of cellular functions, such as RNA processing, RNA export, regulation of chromosome structure, cell cycle progression, initiation of replication, microtubule structure, etc. (for review see Dasso, 1993; Sazer, 1996). One could argue that these effects are all secondary consequences from an impaired NLS-dependent protein import. However, it is also possible that these defects are more direct and that eukaryotic cells contain many immediate targets of Ran function.
Here we describe a novel superfamily of Ran-binding proteins, which includes about a dozen factors in yeast and probably even more in higher eukaryotes. The members of this superfamily share with importin-β an NH2-terminal sequence motif that appears to account for RanGTP binding. Indeed we could confirm the interaction with Ran for the following factors: RanBP7 and RanBP8, two novel, related proteins described here, Cse1p, a cell cycle regulator in yeast, CAS, which is required for apoptosis in cultured human cells, and for Msn5p and Pse1p from yeast. Of these we have characterized RanBP7 and RanBP8 in more detail. Both resemble closely importin-β in their interaction with Ran, and both bind directly to nuclear pore complexes. RanBP7 can cross the nuclear membrane rapidly and in both directions. We provide evidence for a transport cycle in which RanBP7 first enters the nucleus, binds RanGTP inside the nucleus as a prerequisite for rapid re-export to the cytoplasm, after which the RanBP7/RanGTP complex becomes finally disassembled by the concerted action of RanBP1 and RanGAP1 in the cytoplasm. We propose that during these transport cycles, RanBP7 would normally carry an as yet unidentified cargo. This means, RanBP7 and possibly also the other members of the RanBP7/Cse1p/ importin-β superfamily could function as transport receptors that shuttle between nucleus and cytoplasm. RanBP7 and importin-β form an abundant, heterodimeric complex in the cytoplasm that appears to have a function different from nuclear import of proteins with a classical NLS. It might be a way to regulate either RanBP7 or importin-β function. Alternatively, the RanBP7/importin-β complex might constitute an import receptor with a substrate specificity different from that of the importin-α/β complex.
Materials and Methods
Purification of RanBP7 and Binding Assays
For small scale purification, 100 μl of a Xenopus egg extract (plus energy-regenerating system) was diluted 10-fold in binding buffer (50 mM Tris/ HCl, pH 7.5, 200 mM NaCl, 10 mM magnesium acetate), and a post-ribosomal supernatant was prepared. This was rotated overnight with 20 μl IgG Sepharose to which a z-tagged IBB domain (Görlich et al., 1996a ) had been prebound. The nonbound fraction was removed, and the beads were washed six times in binding buffer plus 0.005% digitonin, including a 1-h wash step. Bound proteins were eluted with 1 M magnesium chloride plus 50 mM Tris/HCl, pH 7.5, and precipitated with 90% isopropanol (final). For sequence analysis, the purification was scaled up 50 times and performed in a column mode. The bound proteins were separated on an SDS gel and visualized by Coomassie staining. The RanBP7 band was excised and digested with trypsin. Peptides were separated by HPLC, and several peaks were sequenced. Analysis of human RanBP7 was essentially the same, except that a 50 μg/ml digitonin extract from HeLa cells (for small scale) or a HeLa extract prepared by mild sonication (large scale) was used as the starting material. The binding to the BSA-NLS conjugate has been described previously (Görlich et al., 1996a ). The other analytical binding assays were performed analogous to the small-scale binding described above. Material bound to z–importin-β or z-RanBP7 was eluted with SDS instead of magnesium chloride. Additional modifications are described in the legends.
Antibodies
Antibodies were raised in rabbits against recombinant Xenopus RanBP7. Affinity purification was on sulfo-Link (Pierce, Rockford, IL) to which RanBP7 had been coupled. The anti-RanBP7 antibody is monospecific on blots with total Xenopus egg extract (not shown). Antibodies against importin-α, importin-β, and Ran have been described before (Görlich et al., 1995 b ).
Molecular Cloning
Based on the partial peptide sequences obtained from RanBP7, two degenerate oligonucleotides AA(AG)ATGAT(TCA)GA(AG)AA(AG)CA (TC)GG and AA(AG)GCNAT(TCA)TT(TC)CA(AG)AA(AG) were synthesized and used to identify full length cDNA clones in a Xenopus oocyte cDNA library. For expression and in vitro translation, RanBP7 was cloned into the pQE70 (Qiagen, Hilden, Germany), zz70, and T7-70 vectors (see below). Comparison of the Xenopus RanBP7 sequence with the GenBank/EMBL/DDBJ database identified a similar cDNA-clone (IMAGE Consortium clone ID34250; ACC 360021; Lennon et al., 1996) that was subsequently shown to code for the NH2 terminus of human RanBP8. The sequence information of the complete coding region of RanBP8 was obtained by 3′ race (Primer: GGATGAAGAGCTGTGGCAAGAAGATCC and Oligo dT18) using KlenTaq enzyme (Clontech Laboratories, Palo Alto, CA) and a human cDNA as a template. The human cDNA was prepared from HeLa cell mRNA using the direct mRNA kit (OligoTex; Qiagen) and a cDNA synthesis kit (Great Length cDNA Kit; Clontech Laboratories).
The complete coding sequence of human RanBP8, human CAS, and human transportin were amplified by PCR from HeLa cDNA and cloned into the NcoI/BamH1 sites of pQE60 or zz60. The zz60 vector was generated by an in-frame insertion into the NcoI site of pQE60 of a BspHI/ NcoI fragment containing two z domains from protein A. The DNAs encoding Cse1p, Pse1p, Pdr6p, or the NH2-terminal 682 amino acids of Msn5p were obtained by PCR amplification from genomic Saccharomyces cerevisiae DNA, using primers containing the appropriate restriction sites for subsequent cloning into expression vectors. Cse1p was cloned into the NcoI/BamHI sites of z60, Pdr6p, and the Msn5p fragment, and the Pse1p fragments were cloned into SphI/BamHI sites of zz70 (a pQE70 derivative in which two z domains are fused in front of the multi-cloning site).
Sequence Analysis and Homology Searches
RanBP7 was subjected to a variety of sequence analysis tools (Bork and Gibson, 1996). Database searches with the Blast series of programs (Altschul et al., 1994) revealed significant similarity with RanBP8 (described in this paper), Nmd5p, D9509.15p, HRC1004p (an open reading frame from S. cerevisiae), CAS (Brinkmann et al., 1995), Cse1p (Xiao et al., 1993), and Lph2p. These were aligned using Clustal W (Thompson et al., 1994) and Macaw (Schuler et al., 1991). The most significant blocks accumulated in the first 150 residues of these proteins. To identify more distant members of this new family, iterative profile searches (Birney et al., 1996; Bork and Gibson, 1996) were performed based on an alignment of the conserved NH2-terminal region. New members of this emerging family were accepted and incorporated into the profile for the next iteration when (a) their scores were higher than 5,000, (b) they were clearly separated from the first false positive, and (c) when the profile matched the NH2 terminus of the respective sequence. Each newly identified member was subjected to reciprocal database searches (Bork and Gibson, 1996). This procedure left several putative homologues in the “twilight zone” of the profile search method applied (Birney et al., 1996) and the superfamily described here might be even larger.
The sequence data for the following aligned proteins are available from GenBank/EMBL/DDBJ under accession numbers given: Xenopus RanBP7, U71082; human RanBP8, U77494; human CAS, U33268; S. cerevisiae Cse1p, P33307; S. cerevisiae Lph2p, U43503; S. cerevisiae Nmd5p, P46970; S. cerevisiae D9509.15p, U32274; S. cerevisiae Yrb4p, P40069; human importin-β, L38951; S. cerevisiae importin-β, U19028; S. cerevisiae HRC1004, S53939; human RanBP5, Y08890; S. cerevisiae Pse1p, P32337; S. cerevisiae Crm1p, P30822; S. cerevisiae Mtr10p, U55020; C. elegans C49h3.7, U42436; S. cerevisiae Los1p, P33418; S. cerevisiae Msn5p, X93302; C. elegans T16g12.3, Z30317; S. cerevisiae Pdr6p, P32767, Schizosaccharomyces pombe Crm1p, p14068, S. pombe HRC1004, z69730.
In Vitro Translation
Importin-β and human transportin were cloned into the T7-60 vector, Xenopus RanBP7 into T7-70. T7-60 and -70 are pQE60 and pQE70 derivatives, respectively, in which the promoter/RBS elements were replaced by a T7 promoter and the 5′ untranslated region from Xenopus β globin to enhance translation efficiency. Labeled proteins were generated by coupled transcription/translation in the reticulocyte TNT system (Promega Biotech, Madison, WI) for 2 h at 30°C.
Oocyte Injections
To study nuclear export, RanBP7 was translated in vitro and injected into nuclei of Xenopus oocytes as described (Kambach and Mattaj, 1992). Two internal injection controls for nuclear integrity were used: the hemoglobin from the reticulocyte lysate, and 14C-labeled BSA (Amersham Buchler, Braunschweig, Germany) which was coinjected. The distribution of endogenous proteins in total oocytes, nuclear, and cytoplasmic fractions was determined by SDS-PAGE followed by Coomassie staining or Western blotting.
Microinjection of RNA and analysis by denaturing gel electrophoresis and autoradiography were performed as described (Jarmolowski et al., 1994). The concentration of the recombinant importin-β and RanBP7 in the injected samples was ∼40 μM. The mutant RNAs used (U1ΔSm, U5ΔSm) lack protein binding sites required for the nuclear reimport of these RNAs and thus remain in the cytoplasm after export from the nucleus. U6Δss RNA is not exported and is used as an internal control for nuclear injection.
Expression and Purification of Recombinant Proteins
The expression and purification of the following factors was as described previously: importin-β, nucleoplasmin core domain, z-tagged IBB domain from Xenopus importin-α (Görlich et al., 1996a ), NTF2 (Kutay et al., 1997), Ran, RanBP1, and S. pombe Rna1p (Bischoff et al., 1995a , b ). z-Tagged RanBP1 was expressed from the zz60 vector and purified via a COOH-terminal his-tag. RanQ69L was expressed from pQE32 (Qiagen) and purified on nickel-NTA agarose (Qiagen), and after nucleotide exchange for GTP, RanQ69L was concentrated on SP Sepharose FF (Pharmacia, Uppsala, Sweden). For transport studies, S. pombe Rna1p was expressed from pQE60 or zz60, and its enzymatic activity was verified by a GAP assay. For some experiments, importin-β was expressed with an NH2-terminal z-tag and purified on an IBB domain coupled to sulfoLink, followed by gel filtration on Superdex 200 (Pharmacia). RanBP7 and RanBP8 were expressed with COOH-terminal his-tags from the pQE70 and pQE60 vectors, respectively. For some experiments two z-domains were fused to their NH2 termini. Expression of RanBP7 and RanBP8 was in the BLR/ Rep4 strain at 15°C, and purification was on nickel agarose followed by chromatography on MonoQ. Pse1p, Cse1p, CAS, Msn5p, and Pdr6p were expressed with NH2-terminal z-tags.
Transportin was expressed at 15°C with a COOH-terminal his-tag, with or without an NH2-terminal z-tag. Purification was with nickel agarose followed by chromatography on MonoQ (Pharmacia). Activity was verified in an import assay using a fluorescent nucleoplasmin core-M9 fusion as a substrate.
Fluorescence Labeling
Labeling of importin-β has been described before (Kutay et al., 1997). RanBP7 and transportin were labeled with fluorescein 5′ maleimide as follows: the recombinant proteins were purified on nickel agarose and MonoQ, omitting DTT and mercaptoethanol during the MonoQ step. The protein concentration was estimated from the UV absorbance at 280 nm (Edelhoch, 1967) and fluorescein 5′ maleimide, dissolved in dimethylformamide, and was added at a 1:1 molar ratio. The reaction was allowed to proceed for 4 h on ice, quenched with 50 mM mercaptoethanol, and the labeled proteins were purified on a Superdex 200 HR column equilibrated in 20 mM Hepes/KOH, 80 mM potassium acetate, 3 mM magnesium acetate. Human IgG was labeled with Fluos (Boehringer Mannheim, Mannheim, Germany) and purified on Superdex 200.
NPC Binding Assay
The binding assay was essentially performed as an import assay with permeabilized cells as described earlier (Görlich et al., 1996a , b ). The binding buffer contained 20 mM Hepes/KOH, pH 7.5, 140 mM potassium acetate, 4 mM magnesium acetate, 1 mM DTT, 250 mM sucrose, and 2 mg/ml nucleoplasmin core, 2 mg/ml BSA, plus 1 mg/ml aprotinin to block nonspecific binding, 1.5 μM Ran (GDP form), 150 nM RanBP1, 150 nM Rna1p, 150 nM NTF2, and an energy-regenerating system containing 0.5 mM GTP and ATP, 10 mM creatine phosphate, and 50 μg/ml creatine kinase. Binding to the NPC was allowed for 10 min at room temperature. The other details are given in the legend. The nuclei were fixed with paraformaldehyde/glutaraldehyde, spun onto coverslips, and analyzed by confocal microscopy.
Enzymatic Assays
Labeling of Ran with γ-[32P]GTP, GTPase, and nucleotide exchange assays were performed as described (Bischoff et al., 1994, 1995b ; Görlich et al., 1996b ; Kutay et al., 1997). The reactions were performed in 20 mM Hepes/KOH, pH 7.4, 100 mM NaCl, 1 mM MgCl2, 1 mM sodium azide, 0.05% hydrolyzed gelatin.
Overlay Blots
The basic method followed a published protocol (Lounsbury et al., 1994, 1996) with some modifications. Briefly, proteins were separated by SDS-PAGE and transferred onto nitrocellulose. The blot was then incubated for 1 h at 4°C in renaturation buffer containing 20 mM MOPS, pH 7.1, 100 mM sodium acetate, 5 mM magnesium acetate, 5 mM dithiothreitol, 0.5% bovine serum albumin, 0.05% Tween 20, and subsequently for 30 min at 25°C in renaturation buffer plus 100 μM unlabeled GTP (10 ml total volume). After 10 min, 100 μl of 1 nM Ran γ-[32P]GTP was added alone or with either 5 μM RanBP1 or importin-β. After a further 10 min the blot was washed five times in renaturation buffer and subjected to autoradiography at −70°C. Overlays with Gsp1p were performed the same way as with Ran.
Results
Identification of RanBP7 as Copurifying with Importin-β
The IBB domain is that part of importin-α to which the β subunit binds with high affinity. At high salt concentrations, importin-β appears to bind from a complete cytosol as a single polypeptide to an immobilized IBB domain (Görlich et al., 1996a , b ). At more physiological ionic strength (200 mM), however, a second band of 120 kD copurifies reproducibly (Fig. 1 A, lane 2). The binding of importin-β and the 120-kD protein to the IBB domain is highly specific by a number of criteria. First, binding requires a functional IBB domain (not shown). Second, the binding of both polypeptides is sensitive to RanGTP. RanGTP binds to importin-β and thereby prevents interaction with importin-α or the IBB domain. We used here the GTPase-deficient RanQ69L mutant which remains in its GTP-bound form, even in the presence of the cytoplasmic RanGAP1 (Klebe et al., 1995). If 10 μM RanQ69L GTP was added to the starting material, neither importin-β nor the 120-kD protein was recovered in the bound fraction (Fig. 1 A, lane 3). Third, binding is competed by excess of the nonimmobilized IBB domain (Fig. 1 B, lane 4). For reasons detailed below, we will refer to the 120-kD protein as Ran binding protein 7 (RanBP7). To facilitate further analysis, we obtained partial peptide sequences from RanBP7, used this information to clone the corresponding gene from a Xenopus oocyte cDNA library, and expressed RanBP7 in Escherichia coli. The RanBP7 sequence and its homologies to other proteins are discussed below.
Figure 1.

A 120-kD Ran-binding protein copurifies with importin-β. (A) A Xenopus egg extract (high speed supernatant, lane 1) was subjected to binding to an immobilized IBB domain at 200 mM NaCl. The starting material (lane 1) and the bound fractions (lanes 2–4) were analyzed by SDS-PAGE followed by Coomassie staining. Two bands are specifically recovered in the IBB-bound fraction, importin-β, and a copurifying 120-kD protein that is referred to here as RanBP7. Note, the addition of 10 μM RanQ69L GTP or nonimmobilized IBB competitor (50 μM) prevent binding of both importin-β and RanBP7 (lanes 3 and 4, respectively). (B) RanBP7 was expressed in E. coli. A lysate was prepared and subjected to binding to an IBB column or to an IBB column to which importin-β had been prebound. The starting material and the bound fractions were analyzed by SDS-PAGE followed by Coomassie staining. Note, binding of RanBP7 to the IBB domain is via importin-β. (C) A digitonin (50 μg/ml) extract from HeLa cells was prepared and bound to an immobilized IBB domain as in A. Analysis was by Coomassie staining and by immunoblotting with affinity purified antibodies raised against RanBP7, importin-β, importin-α (Rch1p), and Ran. Again, two bands were recovered specifically in the bound fraction: importin-β and a 120-kD protein that cross-reacts with the anti–Xenopus RanBP7 antibody. (D) A blot of HeLa cell extract and the corresponding IBB-bound fraction (as in C) was probed with a RanBP1/Ran γ-[32P]GTP complex, followed by autoradiography. In the IBB-bound fraction, both importin-β and RanBP7 give a strong signal. In the crude lysate, importin-β and a double band of 120 kD is detected. The latter consists probably of several Ran-binding proteins, in addition to RanBP7.
The finding that RanBP7 copurifies with importin-β on an IBB column can be explained in two ways. First, importin-β and RanBP7 could each bind independently to the IBB domain, or, secondly, RanBP7 could bind via importin-β. To distinguish between these possibilities, we subjected a lysate from E. coli expressing RanBP7 to binding to an IBB column. As seen from Fig. 1 B, RanBP7 was recovered in the bound fraction only when importin-β was added. Thus, RanBP7 binds to the IBB domain via importin-β and is therefore an importin-β binding protein.
To find out whether the expression of RanBP7 is restricted to Xenopus oocytes or also common to other tissues, we raised antibodies against recombinant RanBP7 and found them to cross-react with a 120-kD protein in a variety of cells, including HeLa cells. Furthermore, the complex of RanBP7 and importin-β can be purified on an immobilized IBB domain from a HeLa cell extract in the same way as from an egg extract (Fig. 1 C). The identity of the copurifying 120-kD polypeptide was verified in two ways: by Western blotting using affinity purified anti-RanBP7 antibodies and by peptide sequencing of the 120-kD band, where all 52 residues of obtained partial sequence matched human RanBP7 (not shown).
RanBP7 Is a Novel Ran-Binding Protein
Mammalian cells contain a number of Ran-binding proteins that can be detected by overlay blots using Ran loaded with [32P]GTP as a probe (Coutavas et al., 1993; Lounsbury et al., 1994, 1996). These Ran-binding proteins fall into two classes. The first class includes those containing a RanBP1 homology domain whose Ran binding is competed by RanBP1, i.e., RanBP1 itself (23 kD; Coutavas et al., 1993), and RanBP2 (358 kD; Wu et al., 1995; Yokoyama et al., 1995). In contrast, the signals of the second class comprising 90- and 120-kD bands cannot be competed with RanBP1. The signal is even further enhanced when the blot is probed with the RanBP1/RanGTP complex (Lounsbury et al., 1996; Kutay et al., 1997). The 90-kD band corresponds to importin-β and is recovered in the IBB-bound fraction (Fig. 1 D). The 120-kD protein which copurifies with importin-β, also gives a significant signal in the overlay. It is therefore RanGTP binding and referred to as Ran binding protein 7.
It should be noted that the 120-kD band in the total HeLa extract actually represents a mixture of different Ran-binding proteins (RanBPs) of similar size. Besides RanBP7, it includes at least one additional protein called RanBP5 (Deane et al., 1997) and probably also RanBP8 (see below). Further candidates are RanBP4 and RanBP6 (Hartmann, E., unpublished observations), which are human homologues to the S. cerevisiae proteins Yrb4p (Schlenstedt, G., E. Smirnova, R. Deane, J. Solsbacher, U. Kutay, D. Görlich, H. Ponstingl, and F.R. Bischoff, manuscript submitted for publication) and Pse1p, respectively. However, of these, only RanBP7 appears to be recovered in the IBB-bound fraction, as judged by microsequencing of the 120-kD band (not shown).
The RanBP7–Importin-β Interaction Appears Unrelated to NLS-mediated Protein Import
Because importin-β is a key mediator of NLS-mediated nuclear protein import, we wanted to know if it interacts with RanBP7 to promote this process. In this case, a quaternary NLS/importin-α/β/RanBP7 complex should be the active species. However, the experiment shown in Fig. 2 A makes this assumption unlikely: only importin-α and -β, but hardly any RanBP7, copurify on an immobilized BSA-NLS conjugate. Consistent with that, RanBP7 does not stimulate import of nucleoplasmin into nuclei of permeabilized cells (not shown). One possibility would be that importin-β binds RanBP7 to modify its substrate specificity. This could then allow the transport of cargoes that carry signals other than a classical NLS. Alternatively, the heterodimer might form to regulate importin-β and/or RanBP7 function.
Figure 2.
The RanBP7–importin-β interaction is regulated by Ran and appears unrelated to NLS-mediated nuclear protein import. (A) A Xenopus egg extract (high speed supernatant) was bound either to an immobilized BSA–NLS conjugate or an IBB domain. The starting material and the bound fractions were analyzed by immunoblots against RanBP7, importin-β, and -α. Note, whereas equal amounts of importin-β were found in both bound fractions, RanBP7 was recovered only with IBB column but hardly at all with the NLS conjugate. (B) z-Tagged RanBP7 was prebound to IgG Sepharose and used to bind importin-β out of a Xenopus egg extract (z is an IgG binding domain from Straphylococcus aureus protein A). Binding was with or without the addition of 10 μM RanQ69L GTP. Starting material and bound fractions were analyzed by immunoblotting against Ran and importin-β, but the z-tag is also detected by this procedure. Note that importin-β but no Ran bound to RanBP7 in the absence of RanQ69L. If however, the Ran mutant was added, importin-β binding was abolished, and instead, Ran (Q69L) was recovered in the bound fraction. (C) RanBP7 was expressed in E. coli. A lysate was prepared and subjected to binding to z-tagged importin-β that had been preabsorbed to IgG Sepharose. Elution from the beads was either with SDS, which also releases z–importin-β from the column, or with RanQ69L GTP which dissociates RanBP7 from importin-β. *, dimerized Ran; **, IgG light chain.
The RanBP7/Importin-β Complex Is Dissociated by RanGTP
The RanBP7/importin-β heterodimer can be formed by binding importin-β from a Xenopus egg extract to z-tagged RanBP7 (Fig. 2 B). In this case, binding of Ran is not observed, probably because the extract contains Ran only in its GDP-bound form for which RanBP7 has no detectable affinity (see below). If RanQ69L GTP that is stabilized in the GTP-bound form is added to the starting material, then two effects are observed. First, Ran is now recovered in the RanBP7-bound fraction, confirming RanBP7 as a RanGTP-binding protein. Second, the formation of the RanBP7/importin-β complex is prevented.
To show that RanGTP can also dissociate a preformed RanBP7/importin-β complex, we performed the experiment shown in Fig. 2 C. RanBP7 was expressed in E. coli and bound to importin-β that was immobilized on IgG Sepharose via a z-tag. SDS elutes both RanBP7 and z–importin-β from the column. When the elution was performed with RanGTP, z–importin-β remained matrix bound, however RanBP7 was dissociated from importin-β and recovered in the eluate (Fig. 2 C). Thus, the RanBP7/importin-β complex does not form in the presence of RanGTP. Furthermore, a preformed complex is dissociated by RanGTP addition.
Sequence and Homologies of RanBP7
The Xenopus RanBP7 cDNA codes for a protein of 119 kD. The peptide sequences from HeLa RanBP7 and a human RanBP7 sequence tag (Acc R49703) indicate that human RanBP7 is ∼95% identical to the Xenopus protein. On the basis of an expressed sequence tag we have also cloned another relative, RanBP8, which is a human protein with 61% identity to RanBP7 (Fig. 3).
Figure 3.
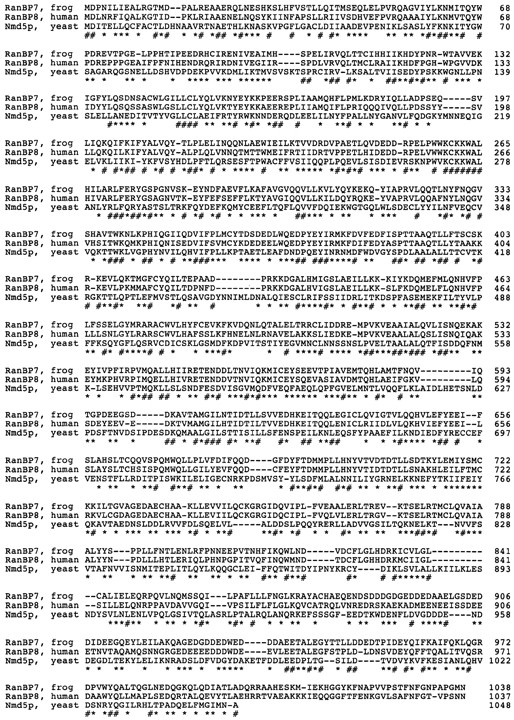
Molecular cloning of Xenopus RanBP7 and human RanBP8 and their similarities to S. cerevisiae Nmd5p. Xenopus RanBP7 was cloned from an oocyte library using partial peptide sequence information from the purified protein. RanBP8 was found in the data base as an expressed human sequence tag similar to RanBP7 and was subsequently cloned from HeLa cDNA. Shown are the aligned amino acid sequences from Xenopus RanBP7, human RanBP8, and S. cerevisiae Nmd5p. Residues identical in all three proteins are indicated by #, and similar amino acids by *. The sequence data for Xenopus RanBP7 and human RanBP8 are available from GenBank/EMBL/DDBJ under accession numbers U71082 and U77494, respectively.
Database searches with the Blast series of programs (Altschul et al., 1994) revealed significant similarity of RanBP7 and RanBP8 to S. cerevisiae Nmd5p (26% identity; Fig. 3), S. cerevisiae D9509.15p, and a number of further proteins of similar length, i.e., 95–130 kD. In the multiple alignment of these sequences (Schuler et al., 1991; Thompson et al., 1994) the most significant blocks were found within the NH2-terminal 150 residues of each protein.
RanBP7 Shares a Common NH2-terminal Sequence Motif with a Protein Superfamily that Includes Importin-β
To identify more distant members of this new family, iterative profile searches (Birney et al., 1996; Bork and Gibson, 1996) were performed based on an alignment of the conserved NH2-terminal region (for details see Materials and Methods section). The results of the procedure are shown in Fig. 4 A. They included more than 20 sequences from different eukaryotes, such as human and yeast importin-β, S. cerevisiae Cse1p, human CAS, S. cerevisiae Crm1p, Nmd5p, etc. This protein superfamily consists of several subgroups, as indicated in Fig. 4 B. The cellular functions attributed to these proteins appear very diverse, but it is possible that all relate to nuclear function. For example, importin-β is the key mediator of NLS-dependent nuclear protein import, while Cse1p has been shown to be required for the destruction of B-type cyclins in the yeast nucleus (Irniger et al., 1995), and human CAS appears to be required for cell proliferation and apoptosis (Brinkmann et al., 1995). Mtr10 was found in a genetic screen for mutants defective in mRNA export from the yeast nucleus (Kadowaki et al., 1994). We reasoned that the common sequence motif in these proteins might relate to a common function.
Figure 4.

The RanBP7/importin-β/Cse1p superfamily. (A) Multiple alignment of amino acids 6–141 from RanBP7 with the NH2 termini of other proteins that have a similar NH2-terminal sequence motif. Positions in red match the consensus. The consensus was defined by the two most frequent amino acids in each position. Proteins named in blue have been shown to bind Ran (see main text and Fig. 5). S. pombe Crm1p and HRC1004 also match the motif but are not shown in the figure. (For accession numbers see Materials and Methods.) (B) Relationship between the proteins shown in A. The tree was calculated from the entire coding sequences and not just from the NH2 termini. Proteins named in red have been shown to bind RanGTP (see also below).
The Conserved NH2-terminal Motif Appears to Account for Interaction with RanGTP
The region of high similarity comprises residues 1–150 in importin-β. We recently mapped the Ran binding domain in importin-β to the NH2-terminal 364 residues (Kutay et al., 1997). The NH2-terminal region from the profile search appears to constitute the conserved core of this domain. That the experimentally found Ran binding domain is larger than the stretch of conserved amino acids can be explained with the need for folding to bring the key residues into the proper spatial position. The conservation profile suggests that these proteins might be targets of Ran function. In this case one would not only predict that proteins like Pse1p and Cse1p bind Ran (or Gsp1p, the yeast Ran, respectively; Belhumeur et al., 1993) but also that the interaction is mediated by the NH2 termini of these proteins. To test the predictions for at least some of the candidates, we expressed human CAS and the yeast proteins Pse1p, Cse1p, Msn5p, and Pdr6p in E. coli, separated the lysates by SDS-PAGE, transferred the proteins onto nitrocellulose, and probed for Ran interaction using Ran (or yeast Gsp1p) charged with labeled GTP as a probe.
The first protein tested for Ran binding is human CAS. We probed this blot with Ran γ-[32P]GTP in the presence of a 5,000-fold excess of RanBP1. This procedure detects Ran-binding proteins of the importin-β type but not Ran binding proteins like RanBP1 or RanBP2. As seen from Fig. 5 A, a control lysate expressing importin-α which does not bind Ran, is negative, but the recombinantly expressed CAS protein gives a positive signal, just as importin-β. The CAS–RanGTP interaction is strongly competed by importin-β (not shown). Thus, human CAS binds RanGTP in a similar way as importin-β.
Figure 5.
Human CAS and the yeast proteins Pse1p, Cse1p, and Msn5p are Ran (Gsp1p) binding. Indicated proteins were expressed in E. coli, and the total lysates were separated by SDS-PAGE and transferred onto nitrocellulose. The blot in A was probed with 10 pM Ran γ-[32P]GTP in the presence of a 5,000-fold molar excess of RanBP1, followed by autoradiography. The negative control was a lysate from E. coli expressing importin-α. The blots in B were probed with 10 pM Gsp1p γ-[32P]GTP (Gsp1p is Ran from S. cerevisiae). The negative control was importin-α, as in A. Pse1p 1–257 and Pse1p 1–409 are fragments of Pse1p comprising of the NH2-terminal 257 or 409 residues. Msn5p was expressed as an NH2-terminal fragment comprised of the NH2-terminal 682 residues.
We next tested Ran binding to full length Pse1p (Chow et al., 1992) and NH2-terminal fragments of Pse1p from S. cerevisiae using γ-[32P]GTP loaded Gsp1p, the Ran homologue of S. cerevisiae. Fig. 5 B shows that full length Pse1p and its 409 NH2-terminal amino acids are positive in this Ran-binding assay. The binding is specific, as it is only observed with the GTP form of Gsp1p and is competed by yeast importin-β (not shown). The NH2-terminal 257 residues, however, are not sufficient to give a significant signal. Thus, the Gsp1 (Ran)-binding domain of yeast Pse1p maps approximately as the Ran binding domain of importin-β (the NH2-terminal 364 amino acids are required for Ran binding). The same figure also demonstrates the Gsp1p binding activity of S. cerevisiae Cse1p (Xiao et al., 1993) and Msn5p (ACC X93302), whereas Pdr6p (Chen et al., 1991), which also came up in the profile search (Fig. 4), and the control lysates gave no signal.
Effects of RanBP7 and RanBP8 on the RanGTPase
Having identified an entire class of novel targets for Ran function, we wanted to investigate the interaction of a representative of this class with Ran in more detail. Ran can cycle between a GDP and GTP-bound state, where the intrinsic rates of nucleotide exchange and GTP hydrolysis are very low. Catalysis by specific factors can increase these rates up to 105-fold (Bischoff and Ponstingl, 1991a ; Bischoff et al., 1994, 1995a ; Becker et al., 1995; Klebe et al., 1995). The major GTPase activating protein is cytoplasmic RanGAP1 (Rna1p in yeast); the principal GTP/GDP exchange factor is RCC1.
Fig. 6 A shows that RanBP7 and RanBP8 inhibit, like importin-β (Floer and Blobel, 1996; Görlich et al., 1996b ), the GAP stimulation of the Ran GTPase. From the dose dependence one can estimate the affinities of these factors for RanGTP. The dissociation constant from RanGTP is ∼25 nM for RanBP7, 4 nM for RanBP8, and 0.8 nM for importin-β, measured under identical conditions. It should be noted that RanBP7 and RanBP8 also inhibit the intrinsic GTPase activity of Ran (not shown).
Figure 6.
Enzymatic interactions of RanBP7 and RanBP8 with Ran. (A) RanBP7 and RanBP8 inhibit GAP induction of GTPase activity of Ran. 50 pM Ran γ-[32P]GTP was preincubated for 30 min with the indicated concentrations of importin-β, RanBP7, and RanBP8. 5 nM Rna1p (RanGAP from S. pombe) was added, and GTP hydrolysis was allowed for 5 min. Hydrolysis of Ran-bound GTP was measured as released γ-[32P]phosphate. When using a very low Ran concentration as in this experiment, the method can be used to estimate the K D of the Ran-binding proteins for RanGTP. These are ∼0.8, 25, and 3 nM for importin-β, RanBP7, and RanBP8, respectively. (B) Effect of RanBP8 on guanine nucleotide exchange on Ran. 30 pM Ran α-[32P]GDP or 50 pM Ran α-[32P]GTP were preincubated for 30 min with indicated concentrations of RanBP8. The exchange reaction was started by addition of 0.2 mM unlabeled GDP plus 2 nM of the exchange factor RCC1. After 5 min, Ran-bound 32P-labeled nucleotide was measured in a filter binding assay. Note, RanBP8 inhibits nucleotide exchange on RanGTP but not on RanGDP. (C) RanBP7 can form a trimeric RanBP7/RanGTP/RanBP1 complex. z-Tagged RanBP1 was prebound to IgG Sepharose. A RanBP7 lysate was subjected to binding to either immobilized RanBP1 alone or to an immobilized RanBP1/RanGTP complex. Elution was with SDS, which also releases the z-RanBP1 from the IgG Sepharose. Note, RanBP7 does not bind to RanBP1 alone, but it is specifically recovered with the RanBP1/RanGTP complex. *, IgG light chain that leaked from the column. (D) The GAP resistance of the RanBP7/RanGTP and the RanBP8/RanGTP complex is relieved by RanBP1. 100 nM RanGTP was preincubated for 30 min with indicated concentrations of RanBP7 or RanBP8. Buffer or 200 nM RanBP1 were added, followed immediately by 200 nM Rna1p. GTP hydrolysis was allowed for 5 min and measured as in A.
Fig. 6 B shows that RanBP8 inhibits the RCC1-mediated nucleotide exchange on RanGTP (RanBP7 has the same effect, not shown). The effect is specific for the GTP form; RanGDP is not affected. RanBP8 and RanBP7 thus behave like importin-β (Görlich et al., 1996b ) in this assay.
Like importin-β, RanBP7 and RanBP8 can each form a trimeric complex with RanGTP and RanBP1. These complexes are evident by gel filtration (not shown) or by affinity chromatography. For example, Fig. 6 C shows that RanBP7 can bind via RanGTP to immobilized RanBP1. RanBP1 has a striking effect on both the RanBP7/RanGTP and the RanBP8/RanGTP complex in that it relieves their GAP resistance (Fig. 6 D). RanBP1 is thus a cofactor for the disassembly of these complexes. This might well be one in vivo function of RanBP1. It should be noted that the one clear difference between importin-β and RanBP7/8 is that the RanBP1/RanGTP/importin-β complex is still GAP resistant (Görlich et al., 1996b ), and its disassembly requires at least one additional factor (Bischoff, F.R., unpublished results).
RanBP7 Can Shuttle between Nucleus and Cytoplasm
RanBP7 is, at steady state, a predominantly cytoplasmic protein (see below). To test if it can cross the nuclear envelope like importin-β, we translated RanBP7 in vitro in the presence of [35S]methionine and injected the mixture into nuclei of Xenopus oocytes. We used two internal controls for nuclear integrity, the hemoglobin from the reticulocyte lysate, which is not exported and clearly visible in the injected nuclei (not shown), and 14C-labeled BSA (“injection control”). After 5 or 90 min, the oocytes were dissected into nuclear and cytoplasmic fractions, which were analyzed for the presence of the labeled proteins. As seen from Fig. 7 A, RanBP7 is rapidly exported from the nuclei, and a significant proportion reached the cytoplasm already after 5 min. After 90 min the steady state distribution was reached in which 90–95% of RanBP7 is cytoplasmic.
Figure 7.
Characterization of RanBP7 transport between nucleus and cytoplasm. (A) RanBP7 was translated in the reticulocyte system in the presence of [35S]methionine and mixed with [14C]BSA, which served as an injection control. The mixture was then injected into Xenopus laevis oocyte nuclei either alone or together with 40 μM Rna1p, or with Rna1p and 80 μM RanQ69L. Proteins were extracted 5 or 90 min after injection. T, C, and N, indicate proteins extracted from total oocytes or after dissection from cytoplasmic or nuclear fractions, respectively. Proteins were analyzed by SDS-PAGE followed by fluorography. (B) Nuclei of Xenopus oocytes were injected either with buffer (control), or 10 μM Rna1p. Oocytes were dissected 6 h later, and the distribution of endogenous proteins was analyzed by SDS-PAGE followed by Coomassie staining, or by Western blotting with RanBP7 and importin-α antibodies. In the control, RanBP7 and importin-α are predominantly cytoplasmic. Nearly 50% of RanBP7 and ∼20% of importin-α accumulated in the nucleus, after nuclear Rna1p injection that inhibits re-export of RanBP7 and importin-α. *, Hemoglobin from the injection control. Rna1p stays nuclear after nuclear injection as judged by Western blotting (not shown).
We next wanted to know if the nuclear export of RanBP7 is active and dependent on specific factors. As RanBP7 is a Ran-binding protein, we reason that its interaction with Ran might be crucial for its transport across the nuclear envelope. As detailed in the introduction, there is a strikingly asymmetric distribution of the Ran system. The GDP/GTP exchange factor RCC1, which loads Ran with GTP, is exclusively nuclear. Its antagonist, the GTPase-activating protein RanGAP1 is excluded from the nucleoplasm and depletes RanGTP from the cytoplasm. As a consequence, the RanGTP concentration should be high in the nucleus and very low in the cytoplasm. We wanted to know if the breakdown of this RanGTP gradient would affect the nuclear export of RanBP7. To deplete the nuclear RanGTP pool, we injected Rna1p (RanGAP1 from S. pombe) into nuclei of Xenopus oocytes. As seen from Fig. 7 A, this treatment severely reduced the rate of RanBP7 export. Strikingly, export was restored if the Rna1p-resistant RanQ69L mutant (GTP form) was prebound to RanBP7 before nuclear injection. Two conclusions are suggested from this experiment. First, RanBP7 requires nuclear RanGTP for export, and second, RanBP7 is exported out of the nucleus as a complex with RanGTP. This also excludes the possibility that RanBP7 leaves the nucleus by simple diffusion. Because RanGTP dissociates RanBP7 from importin-β (Fig. 2, B and C), we can also exclude that RanBP7 is exported by importin-β.
At a steady state, RanBP7 is 90–95% cytoplasmic, and 5–10% are found in the nuclear fraction (Fig. 7 B, control). If this distribution reflects the dynamics of import and export, then endogenous RanBP7 should shift to the nucleus if its re-export from the nucleus is inhibited by a nuclear injection of Rna1p (S. pombe RanGAP1). As seen from Fig. 7 B this treatment does not change the localization of any of the major cytoplasmic proteins (Fig. 7 B, Coomassie), however, ∼50% of the total RanBP7 are indeed chased into the nucleus. Considering that the nucleus has only 1/10 of the volume of the cytoplasm, this is not just equilibration between nucleus and cytoplasm but rather is a 10-fold accumulation in the nuclei. The treatment also causes an increase in nuclear importin-α. This is consistent with data in S. cerevisiae (Koepp et al., 1996) showing that SRP1p (yeast importin-α) accumulates in the nucleus if Prp20p (yeast nucleotide exchange factor for Ran) is defective, which probably also causes a decrease in the nuclear RanGTP concentration. This would suggest that the export of importin-α and RanBP7 requires a higher nuclear RanGTP concentration than their import.
RanBP7 and RanBP8 Bind to Nuclear Pore Complexes
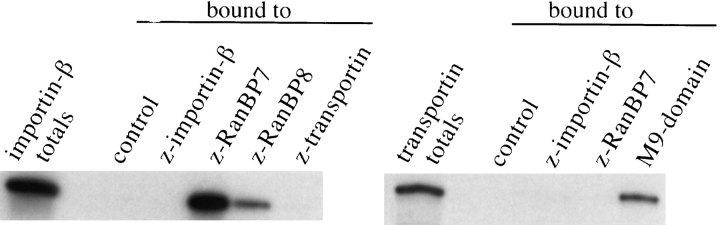
Having shown that RanBP7 can cross the nuclear envelope, we wanted to know whether RanBP7 makes a direct contact to nuclear pores. To test this, permeabilized cells were incubated with z-tagged importin-β (as a positive control) or with z-RanBP7. The binding of the z-tagged proteins was visualized with fluorescent IgG added to the import reaction. Fig. 8 shows the fluorescent staining as recorded with a confocal microscope. Strikingly, when z–importin-β or z-RanBP7 were present, the typical NPC staining pattern was observed.
Figure 8.

RanBP7 and RanBP8 bind to nuclear pore complexes. (A) Permeabilized cells were incubated as indicated with 300 nM z-tagged importin-β, RanBP7, or RanBP8 and with 300 nM or ∼3 μM untagged importin-β. z-Tagged proteins were visualized with 300 nM fluorescein-labeled human IgG added to the import assay. Ran and an energy mix were also added. Shown are confocal sections through the equators of the fixed nuclei. Note, z–importin-β, z-RanBP7, and z-RanBP8 give the typical nuclear pore staining pattern of narrow, punctate rims. NPC binding of RanBP7 and RanBP8 is competed by untagged importin-β, and a fluorescent signal is only observed in the cytoplasmic remnants surrounding the nuclei.
RanBP7 could bind directly to NPCs or only in a complex with importin-β. In the latter case, the RanBP7–NPC interaction should be importin-β dependent. Permeabilized cells still contain endogenous importin-β, however only 10–20% of the available importin-β binding sites at the NPC are occupied by the endogenous protein (Görlich et al., 1995). If RanBP7 binding to the NPC would be strictly importin-β dependent, it should be stimulated by exogenous importin-β. However, NPC binding of z-RanBP7 was not increased but even lower if a 1:1 complex with nontagged importin-β was preformed and completely competed by a 10-fold excess of importin-β. Fig. 8 also shows that RanBP8 is NPC binding, and again, this binding is competed by excess of importin-β.
The data already suggest that RanBP7 binds to NPC not via importin-β but directly and to the same sites as importin-β. However, the interpretation of the competition is somewhat complicated by the fact that at least 3 species have to be considered, RanBP7, importin-β, and the dimeric complex of the two. We therefore wanted to know if a transport receptor with no affinity for RanBP7 would also compete with RanBP7 for NPC binding.
Fig. 9 shows that importin-β does not bind to itself or to transportin but binds strongly to RanBP7 and about four times less efficiently to RanBP8. Transportin, in turn, has no detectable affinity for either importin-β or RanBP7. We therefore chose transportin for the next competition experiment, in which importin-β, transportin, and RanBP7 were each directly fluorescein labeled and subjected at 30 nM to NPC binding. Importin-β was the positive control in this experiment. Transportin also showed a clear NPC binding (it should be noted that transportin accumulates inside the nuclei if added at a higher concentration, probably because it then saturates its own export out of the nucleus). Strikingly, the transportin signal at the nuclear envelope could be competed by unlabeled RanBP7. Fluorescent RanBP7 gave a very clear nuclear pore staining, which could be competed by unlabeled RanBP7 itself or by transportin.
Figure 9.
Interactions between RanBP7, RanBP8, importin-β, and transportin. Importin-β and transportin were translated in a reticulocyte lysate in the presence of [35S]methionine. 200 nM z-tagged importin-β, RanBP7, RanBP8, or transportin were added as indicated, and complexes were allowed to form, which were subsequently recovered with IgG Sepharose. The control binding was without a z-tagged protein. An immobilized M9 domain (the import substrate of transportin) was the positive control for transportin binding.
As a consequence of the competition for NPC binding between the different transport mediators, excess of one transport receptor should compete the transport mediated by another one. Indeed, we have previously shown that dominant-negative importin-β mutants or excess of wild-type importin-β inhibit mRNA and U snRNA export, probably by saturating common binding sites at the nuclear pore complex (Kutay et al., 1997). Fig. 11 shows that RanBP7 also competes mRNA and U snRNA export, most likely at the level of NPC binding. As for importin-β, the export of tRNA is not affected at this concentration.
Figure 11.
mRNA and U snRNA export are competed by importin-β or RanBP7. Xenopus laevis oocyte nuclei were injected with buffer or 40 μM of importin-β or RanBP7 (as indicate above the lanes) together with a mixture of the following radioactively labeled RNAs: DHFR mRNA, histone H4 mRNA, U1ΔSm, U5ΔSm, U6Δss, and human initiator methionyl tRNA. U6Δss does not leave the nucleus and is an internal control for nuclear integrity. Synthesis of DHFR, histone H4, U1ΔSm, and U5ΔSm RNAs was primed with the m7GpppG cap dinucleotide, whereas synthesis of U6Δss RNA was primed with γ-mGTP. RNA was extracted either 5 or 150 min after injection and detected by electrophoresis, followed by autoradiography.
Discussion
The small GTPase Ran has been implicated in a variety of cellular functions, such as transport into and out of the nucleus, regulation of cell cycle progression, and chromatin structure (for reviews see Dasso, 1993; Sazer, 1996). However, with the exception of the importin-dependent transport pathway, no immediate targets of Ran function have been reported so far. We have now identified a novel class of RanGTP binding proteins, which includes to date about 20 proteins from various eukaryotes. The members of this protein superfamily share the following structural features: (a) a common NH2-terminal sequence motif; (b) a similar large size of 90–130 kD; (c) a similar isoelectric point ∼4.6 (ranging from 4.3–5.4); and (d) predicted secondary structures with extensive helical regions. In addition, at least some of them have a clear internal repeat structure (Andrade and Bork, 1995). Whether or not this applies to all members is currently under investigation.
The members of the RanBP7/Cse1p/importin-β proteins superfamily (Fig. 4, and Fornerod et al., 1997) can be grouped roughly into families as follows: (a) importin-β–like proteins including Yrb4p and Pse1p from yeast and RanBP5 from human. (b) A family represented by Los1p and (c) Crm1p from yeast and human. (d) A family including Msn5p, Mtr10p, and HRC1004p, and finally (e) Cse1p-like proteins such as CAS, RanBP7, RanBP8, D95009.15p, and Nmd5p. Except for the NH2-terminal sequence motif, there is not much homology between these groups of proteins.
In importin-β, the conserved NH2-terminal region is essential for RanGTP binding (Görlich et al., 1996b ; Kutay et al., 1997). The presence of this motif in the other members of the superfamily would suggest that they interact with RanGTP as well. This is indeed the case for at least RanBP7, RanBP8, CAS, Pse1p, Msn5p, Cse1p (Figs. 1, 2 B, 5, and 6), RanBP5 (Deane et al., 1997), and Yrb4p (Schlenstedt, G., E. Smirnova, R. Deane, J. Solsbacher, U. Kutay, D. Görlich, H. Ponstingl, and F.R. Bischoff, manuscript submitted for publication). From the tested proteins in Fig. 5, only Pdr6p (Chen et al., 1991) appears negative for Ran interaction. This might indicate a limitation in the assays employed, or it might mark the border line for the significance of the profile in Fig. 4 A. On the other hand, not all Ran-binding proteins of this kind are covered by the consensus in Fig. 4 A. For example, transportin does not match the consensus, although it shows otherwise good overall homology with importin-β, functions similarly as importin-β (Pollard et al., 1996), and also binds Ran (Bischoff, F.R., S. Nakielny, and G. Dreyfuss, unpublished results).
It should be emphasized that the RanBP7/Cse1p/importin-β protein superfamily is distinct from the family of Ran-binding proteins with a RanBP1 homology domain. The two Ran-binding motifs are unrelated in sequence, their effects on the GTPase are different, and their binding sites on Ran are distinct and nonoverlapping (Fig. 6, and Introduction).
The interaction with the nuclear pore complex appears to be a second characteristic of the RanBP7/Cse1p/importin-β superfamily. This has been well established for importin-β and is shown here for RanBP7 and RanBP8. In addition, binding to the NPC or an NPC-like cellular distribution has been demonstrated for RanBP5 (Deane et al., 1997), Yrb4p (Schlenstedt, G., E. Smirnova, R. Deane, J. Solsbacher, U. Kutay, D. Görlich, H. Ponstingl, and F.R. Bischoff, manuscript submitted for publication), human Crm1p (Fornerod et al., 1997), Los1p (Simos et al., 1996), and Cse1p (cited in Irniger et al., 1995). The potential to interact with both Ran and the NPC would suggest that transport across the nuclear envelope might be a common function of this superfamily of proteins. Each of the candidates might represent one distinct transport pathway, and the task for the future will be to identify their cargoes.
We have previously observed that importin-β and importin-β fragments cross-compete with other major nucleocytoplasmic transport pathways such as M9-mediated import and the export of NES containing proteins, mRNA, and U snRNA (Kutay et al., 1997). This supports the view that the mediators of these pathways function similarly to importin-β, probably take a similar path through the NPC, and share at least some of the intermediate binding sites. Here we show that RanBP7 cross-competes with mediators of mRNA and U snRNA export as well. Furthermore, we observed a direct competition for binding to the nuclear pore complex between RanBP7, importin-β, and transportin. Thus, also by this criterion, RanBP7 behaves like a shuttling transport receptor.
RanBP7 and RanBP8 are Ran-binding proteins with similar properties as importin-β. Although the affinity of RanBP7 for RanGTP is lower than that of importin-β, it is still in a physiological range. The K D of the RanBP7/ RanGTP complex is, with ∼25 nM, still far below the cellular Ran concentration of several μM. RanBP7 binds specifically, the GTP-bound form of Ran. As free RanGTP should be available only inside the nucleus, the RanBP7/ RanGTP complex should assemble only in this compartment. Once the RanBP7/RanGTP complex is formed, it is remarkably stable. The offrate is ∼2 h, and the low intrinsic GTPase activity of Ran is even further reduced (not shown). Furthermore, a GTPase activation by RanGAP1 (RNA1p) is prevented, and the bound GTP is protected against nucleotide exchange (Fig. 6).
The disassembly of the RanBP7/RanGTP requires the concerted action of RanBP1 and RanGAP1 (Fig. 6). We suggest the following working model for this process: the intermediate of disassembly is the trimeric RanBP7/RanGTP/ RanBP1 complex (Fig. 6 C), which appears to be in a fast equilibrium with free RanBP7 and the dimeric RanGTP/ RanBP1 complex (Bischoff, F.R., and D. Görlich, manuscript in preparation). The latter is an excellent substrate for RanGAP1, which triggers GTP hydrolysis and thereby makes the disassembly of the complex irreversible. RanBP1 is essential for nuclear transport in yeast (Schlenstedt et al., 1995b ), but at which step it functions had been obscure so far. It is therefore an attractive possibility to assume the physiological function of RanBP1 in the recycling of transport factors like RanBP7.
The RanBP7/RanGTP complex should form only in the nucleus; however, its disassembly is most likely cytoplasmic because the two factors required for disassembly, RanGAP1 and RanBP1, are excluded from the nucleoplasm (Hopper et al., 1990; Matunis et al., 1996; Richards et al., 1996; Mahajan et al., 1997). Assuming that RanBP7 shuttles between nucleus and cytoplasm, one therefore would predict that RanBP7 leaves the nucleus as a complex with RanGTP. In fact, RanBP7 becomes rapidly exported when injected into nuclei of Xenopus oocytes. Already after 5 min, ∼20% of RanBP7 has reached the cytoplasm. Considering the large size of a Xenopus oocyte, this is exceedingly fast, in fact much faster than the export of a BSA–NES conjugate and even faster than tRNA export. The export rate is significantly reduced if nuclear RanGTP is depleted by nuclear injection of Rna1p (RanGAP1). The block is not complete, but this is understandable considering that only 25 nM RanGTP is required for half-maximal binding to RanBP7, compared to at least 10 μM present normally in the nucleus. Thus, to have a significant effect, the nuclear RanGTP concentration probably has to be lowered by three orders of magnitude, counteracting the continuous RanGTP production by RCC1. RanGTP depletion from the nucleus inhibits RanBP7 export probably by preventing the RanBP7/ RanGTP complex formation because the export can be restored by prebinding of RanQ69L GTP (a GAP-resistant Ran mutant) to RanBP7. This further supports the model that RanBP7 is indeed exported as a complex with RanGTP.
RanBP7 is at a steady state predominantly cytoplasmic, and only ∼5% is found in the nuclear fraction. However, this situation becomes strikingly different if nuclear export of RanBP7 is inhibited, and then ∼50% of RanBP7 can be shifted to the nucleus. Considering that the nucleus accounts for only 1/10 of the volume of the oocyte, RanBP7 has become 10-fold concentrated in the nucleus compared to the cytoplasm. This accumulation cannot be explained by simple diffusion. Taken together, RanBP7 can cross the nuclear envelope rapidly and in both directions. Although we have not yet identified a cargo, RanBP7's properties resemble those of a shuttling transport receptor. We do not know if RanBP7 would confer import or export. In analogy to importin-β, however, we would expect the RanGTP– RanBP7 interaction to regulate substrate binding. This would ensure that the substrate is transported in one direction only, even though RanBP7 shuttles.
In Xenopus eggs or HeLa cells, RanBP7 and importin-β form an abundant, cytoplasmic heterodimer. A weaker interaction was found also between RanBP8 and importin-β but so far not between other members of the protein superfamily. A complex between importin-β and an unidentified 120-kD protein has been reported before (Chi et al., 1995), which was probably identical with RanBP7 or RanBP8. The RanBP7/importin-β heterodimer appears not to be involved in the import of proteins with a classical NLS. It might be a way to regulate either RanBP7 or importin-β function, to lower the concentration of free importin-β if there is only a little substrate in the cytoplasm and therefore no need for import. Alternatively, the RanBP7/importin-β complex might constitute an import receptor with a substrate specificity different from that of the importin-α/β complex. Both possibilities are currently being tested.
The members of the RanBP7/Cse1p/importin-β superfamily have been implicated in a variety of cellular functions. For example, mtr10 was identified in a screen for defects in RNA export from the yeast nucleus (Kadowaki et al., 1994). Crm1p is an essential protein in yeast and has been implicated in the maintenance of chromosome structure (Toda et al., 1992; Funabiki et al., 1993). Human CAS appears to be required for cell proliferation, but lowering its cellular levels by the moderate expression of anti-sense RNA renders cultured cells resistant to TNF-triggered apoptosis (Brinkmann et al., 1995). One of the most interesting proteins is, however, S. cerevisiae Cse1p. It is encoded by an essential gene and was originally found in a screen for mutants defective in chromosome segregation (Xiao et al., 1993). Subsequently, it was shown that Cse1p is essential for the destruction of the B-type cyclin CLB2 and thus required for progression through mitosis (Irniger et al., 1995). In yeast, the nuclear envelope does not break down during mitosis and cyclin CLB2 degradation takes place in the nucleus. Our data suggest that Cse1p might be a Ran-binding transport factor that could define a novel nuclear transport pathway. In this model, Cse1p would function in the timely nuclear transport of a regulator or a mediator of cyclin CLB2 degradation. At high copy number, SRP1 (yeast importin-α) can partially suppress the phenotype of the cold-sensitive cse1-1 allele (Xiao et al., 1993), suggesting that some of the Cse1p function can be taken over by the “standard” import pathway. Interestingly, the srp1-31 mutant allele also has a cell cycle phenotype, arresting at nonpermissive temperature in G2/M (Loeb et al., 1995). CLB2 is stabilized also in this strain compared to wild-type cells.
Taken together, yeast might use about a dozen independent nuclear transport pathways. From expressed, sequenced tags in the data base it appears there might be considerably more in higher organisms. This probably enables eukaryotes to make full use of their compartmentalization and to use specific regulated nuclear pathways to control key cellular events.
Figure 10.
Transportin and RanBP7 compete with each other for NPC binding. Importin-β, transportin, and RanBP7 were labeled directly with fluorescein and incubated at a concentration of 50 nM with permeabilized cells in the presence of Ran and an energy-regenerating system. Where indicated, the NPC binding was competed with 2 μM of unlabeled RanBP7 or transportin. For analysis, the samples were fixed, and nuclei were spun onto coverslips and examined by confocal fluorescence microscopy.
Acknowledgments
We wish to thank Susanne Kostka and Regine Kraft for protein sequencing, Petra Schwarzmaier for technical help, Drs. Róisín Deane, Iain W. Mattaj, and Tom A. Rapoport for critical reading of the manuscript. We are grateful to Drs. E. Darzynkiewicz and J. Stepinski for the gift of the 7-methyl GpppG and γ-methyl GTP cap analogues, and Dr. D. Schümperli for the mouse Histone H4 clone.
This work was supported by the Deutsche Forschungsgemeinschaft, the Swiss National Science Foundation (grant 3100-046841), and the State of Geneva. M. Dabrowski is a recipient of a postdoctoral fellowship from the Max-Delbrück-Zentrum für Molekulare Medizin.
Footnotes
1. Abbreviations used in this paper: GAP, GTPase activating protein; IBB, importin β binding; NLS, nuclear localization signal; NPC, nuclear pore complex.
Please address all correspondence to Dirk Görlich, Zentrum für Molekulare Biologie der Universität Heidelberg, Im Neuenheimer Feld 282, 69120 Heidelberg, Germany. Tel.: (49) 6221-546862; Fax: (49) 6221-545892; E-mail: [email protected]
References
- Adam SA, Gerace L. Cytosolic proteins that specifically bind nuclear location signals are receptors for nuclear import. Cell. 1991;66:837–847. doi: 10.1016/0092-8674(91)90431-w. [DOI] [PubMed] [Google Scholar]
- Aitchison JD, Blobel G, Rout MP. Kap104: a karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science (Wash DC) 1996;274:624–627. doi: 10.1126/science.274.5287.624. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Boguski MS, Gish W, Wootton JC. Issues in searching molecular sequence databases. Nat Genet. 1994;6:119–129. doi: 10.1038/ng0294-119. [DOI] [PubMed] [Google Scholar]
- Andrade MA, Bork P. HEAT repeats in the Huntington's disease protein. Nat Genet. 1995;11:115–116. doi: 10.1038/ng1095-115. [DOI] [PubMed] [Google Scholar]
- Becker J, Melchior F, Gerke V, Bischoff FR, Ponstingl H, Wittinghofer A. RNA1 encodes a GTPase-activating protein specific for Gsp1p, the Ran/TC4 homologue of Saccharomyces cerevisiae. . J Biol Chem. 1995;270:11860–11865. doi: 10.1074/jbc.270.20.11860. [DOI] [PubMed] [Google Scholar]
- Beddow AL, Richards SA, Orem NR, Macara IG. The Ran/TC4 GTPase-binding domain: identification by expression by expression cloning and characterizaiton of a conserved sequence motif. Proc Natl Acad Sci USA. 1995;92:3328–3332. doi: 10.1073/pnas.92.8.3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belhumeur P, Lee A, Tam R, DiPaolo T, Fortin N, Clark MW. GSP1 and GSP2, genetic suppressors of the prp20-1 mutant in Saccharomyces cerevisiae: GTP-binding proteins involved in the maintenance of nuclear organization. Mol Cell Biol. 1993;13:2152–2161. doi: 10.1128/mcb.13.4.2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Thompson JD, Gibson TJ. PairWise and SearchWise: finding the optimal alignment in a simultaneous comparison of a protein profile against all DNA translation frames. Nucleic Acids Res. 1996;24:2730–2739. doi: 10.1093/nar/24.14.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff FR, Ponstingl H. Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature (Lond) 1991a;354:80–82. doi: 10.1038/354080a0. [DOI] [PubMed] [Google Scholar]
- Bischoff FR, Ponstingl H. Mitotic regulator protein RCC1 is complexed with a nuclear ras-related polypeptide. Proc Natl Acad Sci USA. 1991b;88:10830–10834. doi: 10.1073/pnas.88.23.10830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff FR, Klebe C, Kretschmer J, Wittinghofer A, Ponstingl H. RanGAP1 induces GTPase activity of nuclear ras-related Ran. Proc Natl Acad Sci USA. 1994;91:2587–2591. doi: 10.1073/pnas.91.7.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff FR, Krebber H, Kempf T, Hermes I, Ponstingl H. Human RanGTPase activating protein RanGAP1 is a homolog of yeast RNA1p involved in messenger RNA processing and transport. Proc Natl Acad Sci USA. 1995a;92:1749–1753. doi: 10.1073/pnas.92.5.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff FR, Krebber H, Smirnova E, Dong WH, Ponstingl H. Coactivation of RanGTPase and inhibition of GTP dissociation by Ran GTP binding protein RanBP1. EMBO J. 1995b;14:705–715. doi: 10.1002/j.1460-2075.1995.tb07049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P, Gibson T. Applying motif and profile searches. Methods Enzymol. 1996;266:162–184. doi: 10.1016/s0076-6879(96)66013-3. [DOI] [PubMed] [Google Scholar]
- Brinkmann U, Brinkmann E, Gallo M, Pastan I. Cloning and characterization of a cellular apoptosis susceptibility gene, the human homologue to the yeast chromosome segregation gene CSE1. Proc Natl Acad Sci USA. 1995;92:10427–10431. doi: 10.1073/pnas.92.22.10427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WN, Balzi E, Capieaux E, Choder M, Goffeau A. The DNA sequencing of the 17 kb HindIII fragment spanning the LEU1 and ATE1 loci on chromosome VII from Saccharomyces cerevisiaereveals the PDR6 gene, a new member of the genetic network controlling pleiotropic drug resistance. Yeast. 1991;7:287–299. doi: 10.1002/yea.320070311. [DOI] [PubMed] [Google Scholar]
- Chi NC, Adam EJH, Adam SA. Sequence and characterization of cytoplasmic nuclear import factor p97. J Cell Biol. 1995;130:265–274. doi: 10.1083/jcb.130.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi NC, Adam EJH, Visser GD, Adam SA. RanBP1 stabilizes the interaction of Ran with p97 in nuclear protein import. J Cell Biol. 1996;135:559–569. doi: 10.1083/jcb.135.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow T, Ash J, Dignard D, Thomas D. Screening and identification of a gene, PSE-1, that affects protein secretion in Saccharomyces cerevisiae. . J Cell Sci. 1992;101:709–719. doi: 10.1242/jcs.101.3.709. [DOI] [PubMed] [Google Scholar]
- Coutavas E, Ren M, Oppenheim JD, D'Eustachio P, Rush MG. Characterization of proteins that interact with the cell-cycle regulatory protein Ran/TC4. Nature (Lond) 1993;366:585–587. doi: 10.1038/366585a0. [DOI] [PubMed] [Google Scholar]
- Dasso M. RCC1 in the cell cycle: the regulator of chromosome condensation takes on new roles. Trends Biochem Sci. 1993;18:96–101. doi: 10.1016/0968-0004(93)90161-f. [DOI] [PubMed] [Google Scholar]
- Deane, R., W. Schäfer, H.-P. Zimmerman, L. Mueller, D. Görlich, S. Prehn, H. Ponstingl, and F.R. Bischoff. 1997. Ran-binding protein 5 (RanBP5) is related to the nuclear transport factor importin-β but interacts differently with RanBP1. Mol. Cell. Biol. In press. [DOI] [PMC free article] [PubMed]
- Dingwall C, Laskey RA. Nuclear targeting sequences—a consensus? . Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- Drivas GT, Shih A, Coutavas E, Rush MG P.D'Eustachio. Characterization of four novel ras-like genes expressed in a human teratocarcinoma cell line. Mol Cell Biol. 1990;10:1793–1798. doi: 10.1128/mcb.10.4.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelhoch H. Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry. 1967;6:1948–1954. doi: 10.1021/bi00859a010. [DOI] [PubMed] [Google Scholar]
- Feldherr CM, Kallenbach E, Schultz N. Movement of karyophilic protein through the nuclear pores of oocytes. J Cell Biol. 1984;99:2216–2222. doi: 10.1083/jcb.99.6.2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floer M, Blobel G. The nuclear transport factor karyopherin β binds stoichiometrically to Ran-GTP and inhibits the Ran GTPase activating protein. J Biol Chem. 1996;271:5313–5316. doi: 10.1074/jbc.271.10.5313. [DOI] [PubMed] [Google Scholar]
- Fornerod M, van Deursen J, van Baal S, Reynolds A, Davis D, Murti KG, Fransen J, Grosveld G. The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. EMBO (Eur Mol Biol Organ) J. 1997;16:807–816. doi: 10.1093/emboj/16.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki H, Hagan I, Uzawa S, Yanagida M. Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J Cell Biol. 1993;121:961–976. doi: 10.1083/jcb.121.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb DS, Gariepy J, Schoolnik G, Kornberg RD. Synthetic peptides as nuclear localization signals. Nature (Lond) 1986;322:641–644. doi: 10.1038/322641a0. [DOI] [PubMed] [Google Scholar]
- Görlich D, Mattaj IW. Nucleocytoplasmic transport. Science (Wash DC) 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- Görlich D, Vogel F, Mills AD, Hartmann E, Laskey RA. Distinct functions for the two importin subunits in nuclear protein import. Nature (Lond) 1995;377:246–248. doi: 10.1038/377246a0. [DOI] [PubMed] [Google Scholar]
- Görlich D, Henklein P, Laskey RA, Hartmann E. A 41 amino acid motif in importin alpha confers binding to importin beta and hence transit into the nucleus. EMBO (Eur Mol Biol Organ) J. 1996a;15:1810–1817. [PMC free article] [PubMed] [Google Scholar]
- Görlich D, Panté N, Kutay U, Aebi U, Bischoff FR. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO (Eur Mol Biol Organ) J. 1996b;15:5584–5594. [PMC free article] [PubMed] [Google Scholar]
- Hopper AK, Traglia HM, Dunst RW. The yeast RNA1 gene product necessary for RNA processing is located in the cytosol and apparently excluded from the nucleus. J Cell Biol. 1990;111:309–321. doi: 10.1083/jcb.111.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamoto N, Shimamoto T, Takao T, Tachibana T, Kose S, Matsubae M, Sekimoto T, Shimonishi Y, Yoneda Y. In vivo evidence for involvement of a 58kDa component of nuclear pore targeting complex in nuclear protein import. EMBO (Eur Mol Biol Organ) J. 1995;14:3617–3626. doi: 10.1002/j.1460-2075.1995.tb00031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irniger S, Piatti S, Michaelis C, Nasmyth K. Genes involved in sister chromatid separation are needed for B-type cyclin proteolysis in budding yeast. Cell. 1995;81:269–278. doi: 10.1016/0092-8674(95)90337-2. [DOI] [PubMed] [Google Scholar]
- Jarmolowski A, Boelens WC, Izaurralde E, Mattaj IW. Nuclear export of different classes of RNA is mediated by specific factors. J Cell Biol. 1994;124:627–635. doi: 10.1083/jcb.124.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki T, Chen SP, Hitomi M, Jacobs E, Kumagai C, Liang S, Schneiter R, Singleton D, Wisniewska J, Tartakoff AM. Isolation and characterization of Saccharomyces cerevisiaemRNA transport-defective mtr mutants. J Cell Biol. 1994;126:649–659. doi: 10.1083/jcb.126.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambach C, Mattaj IW. Intracellular distribution of the U1A protein depends on active transport and nuclear binding to U1 snRNA. J Cell Biol. 1992;118:11–21. doi: 10.1083/jcb.118.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebe C, Bischoff FR, Ponstingl H, Wittinghofer A. Interaction of the nuclear GTP-binding protein Ran with its regulatory proteins RCC1 and RanGAP1. Biochemistry. 1995;34:639–647. doi: 10.1021/bi00002a031. [DOI] [PubMed] [Google Scholar]
- Koepp DM, Silver PA. A GTPase controlling nuclear trafficking: running the right way or walking randomly? . Cell. 1996;87:1–4. doi: 10.1016/s0092-8674(00)81315-x. [DOI] [PubMed] [Google Scholar]
- Koepp D, Wong D, Corbett A, Silver PA. Dynamic localization of the nuclear import receptor and its interactions with transport factors. J Cell Biol. 1996;133:1163–1176. doi: 10.1083/jcb.133.6.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U, Izaurralde E, Bischoff FR, Mattaj IW, Görlich D. Dominant-negative mutants of importin-β block multiple pathways of import and export through the nuclear pore complex. EMBO (Eur Mol Biol Organ) J. 1997;16:1153–1163. doi: 10.1093/emboj/16.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennon GG, Auffray C, Polymeropoulos M, Soares MB. The I.M.A.G.E. Consortium: an integrated molecular analysis of genomes and their expression. Genomics. 1996;33:151–152. doi: 10.1006/geno.1996.0177. [DOI] [PubMed] [Google Scholar]
- Loeb DJ, Schlenstedt G, Pellman D, Kornitzer D, Silver PA, Fink G. The yeast nuclear import receptor is required for mitosis. Proc Natl Acad Sci USA. 1995;92:7647–7651. doi: 10.1073/pnas.92.17.7647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lounsbury KM, Macara IG. Ran-binding protein 1 (RanBP1) forms a ternary complex with Ran and karyopherin β and reduces Ran GTPase-activating Protein (RanGAP) inhibition by karyopherin β. J Biol Chem. 1997;272:551–555. doi: 10.1074/jbc.272.1.551. [DOI] [PubMed] [Google Scholar]
- Lounsbury KM, Beddow AL, Macara IG. A family of proteins that stabilize the Ran/TC4 GTPase in its GTP-bound conformation. J Biol Chem. 1994;269:11285–11290. [PubMed] [Google Scholar]
- Lounsbury KM, Richards SA, Perlungher RR, Macara IG. Ran binding domains promote the interaction of ran with p97/β-karyopherin, linking the docking and translocation steps of nuclear import. J Biol Chem. 1996;271:2357–2360. doi: 10.1074/jbc.271.5.2357. [DOI] [PubMed] [Google Scholar]
- Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- Matunis MJ, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase–activating protein between cytosol and nuclear pore complex. J Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchior F, Paschal B, Evans E, Gerace L. Inhibition of nuclear protein import by nonhydrolyzable analogs of GTP and identification of the small GTPase Ran/TC4 as an essential transport factor. J Cell Biol. 1993;123:1649–1659. doi: 10.1083/jcb.123.6.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael WM, Choi M, Dreyfuss G. A nuclear export signal in hnRNP A1: a signal-mediated, temperature-dependent nuclear protein export pathway. Cell. 1995;83:415–422. doi: 10.1016/0092-8674(95)90119-1. [DOI] [PubMed] [Google Scholar]
- Michaud N, Goldfarb DS. Multiple pathways in nuclear transport: the import of U2 snRNP occurs by a novel kinetic pathway. J Cell Biol. 1991;112:215–223. doi: 10.1083/jcb.112.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MS, Blobel G. The GTP-binding protein Ran/TC4 is required for protein import into the nucleus. Nature (Lond) 1993;365:661–663. doi: 10.1038/365661a0. [DOI] [PubMed] [Google Scholar]
- Moroianu J, Hijikata M, Blobel G, Radu A. Mammalian karyopherin a1b and a2b heterodimers: a1 or a2 subunit binds nuclear localization sequence and b subunit interacts with peptide repeat containing nucleoporins. Proc Natl Acad Sci USA. 1995;92:6532–6536. doi: 10.1073/pnas.92.14.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newmeyer DD, Lucocq JM, Bürglin TR, De Robertis EM. Assembly in vitro of nuclei active in nuclear protein import. EMBO (Eur Mol Biol Organ) J. 1986;5:501–510. doi: 10.1002/j.1460-2075.1986.tb04239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo M, Kai R, Furuno N, Sekiguchi T, Sekiguchi M, Hayashida H, Kuma K-I, Miyata T, Fukushige S, Murotsu T, et al. Isolation and characterization of the active cDNA of the human cell cycle gene RCC1 involved in the regulation of onset of chromosome condensation. Genes Dev. 1987;1:585–593. doi: 10.1101/gad.1.6.585. [DOI] [PubMed] [Google Scholar]
- Ohtsubo M, Okazaki H, Nishimoto T. The RCC1 protein, a regulator for the onset of chromosome condensation locates in the nucleus and binds to DNA. J Cell Biol. 1989;109:1389–1397. doi: 10.1083/jcb.109.4.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panté N, Aebi U. Towards the molecular dissection of protein import into nuclei. Curr Opin Cell Biol. 1996;8:397–406. doi: 10.1016/s0955-0674(96)80016-0. [DOI] [PubMed] [Google Scholar]
- Palacios I, Weis K, Klebe C, Dingwall C. Ran/TC4 mutants identify a common requirement for snRNP and protein import into the nucleus. J Cell Biol. 1996;133:485–494. doi: 10.1083/jcb.133.3.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard VW, Michael WM, Naldalny B, Slomi MC, Wang P, Dreyfuss G. A novel receptor mediated nuclear protein import pathway. Cell. 1996;88:985–994. doi: 10.1016/s0092-8674(00)80173-7. [DOI] [PubMed] [Google Scholar]
- Rexach M, Blobel G. Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell. 1995;83:683–692. doi: 10.1016/0092-8674(95)90181-7. [DOI] [PubMed] [Google Scholar]
- Richards SA, Lounsbury KM, Carey KL, Macara IG. A nuclear export signal is essential for the cytosolic localization of the Ran binding protein, RanBP1. J Cell Biol. 1996;134:1157–1168. doi: 10.1083/jcb.134.5.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sazer S. The search for the primary function of the Ran GTPase continues. Trends Cell Biol. 1996;6:81–85. doi: 10.1016/0962-8924(96)80992-5. [DOI] [PubMed] [Google Scholar]
- Schlenstedt G. Protein import into the nucleus. FEBS (Fed Eur Biochem Sci) Lett. 1996;389:75–79. doi: 10.1016/0014-5793(96)00583-2. [DOI] [PubMed] [Google Scholar]
- Schlenstedt G, Saavedra C, Loeb JDJ, Cole CN, Silver PA. The GTP-bound form of the yeast Ran/TC4 homologue blocks nuclear protein import and appearance of polyA+RNA in the cytoplasm. Proc Natl Acad Sci USA. 1995a;92:225–229. doi: 10.1073/pnas.92.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenstedt G, Wong DH, Koepp DM, Silver PA. Mutants in a yeast Ran binding protein are defective in nuclear transport. EMBO (Eur Mol Biol Organ) J. 1995b;14:5367–5378. doi: 10.1002/j.1460-2075.1995.tb00221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler GD, Altschul SF, Lipman DJ. A workbench for multiple alignment construction and analysis. Proteins. 1991;9:180–190. doi: 10.1002/prot.340090304. [DOI] [PubMed] [Google Scholar]
- Sweet DJ, Gerace L. Taking from the cytoplasm and giving to the pore: soluble transport factors in nuclear protein import. Trends Cell Biol. 1995;5:444–447. doi: 10.1016/s0962-8924(00)89108-4. [DOI] [PubMed] [Google Scholar]
- Simos G, Tekotte H, Grosjean H, Segref A, Sharma K, Tollervey D, Hurt EC. Nuclear pore proteins are involved in the biogenesis of functional tRNA. EMBO (Eur Mol Biol Organ) J. 1996;15:2270–2284. [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T, Shimanuki M, Saka Y, Yamano H, Adachi Y, Shirakawa M, Kyogoku Y, Yanagida M. Fission yeast pap1-dependent transcription is negatively regulated by an essential nuclear protein, crm1. Mol Cell Biol. 1992;12:5474–5484. doi: 10.1128/mcb.12.12.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis K, Mattaj IW, Lamond AI. Identification of hSRP1α as a functional receptor for nuclear localization sequences. Science (Wash DC) 1995;268:1049–1053. doi: 10.1126/science.7754385. [DOI] [PubMed] [Google Scholar]
- Weis K, Dingwall C, Lamond AI. Characterization of the nuclear protein import mechanism using Ran mutants with altered binding specificities. EMBO (Eur Mol Biol Organ) J. 1996a;15:7120–7128. [PMC free article] [PubMed] [Google Scholar]
- Weis K, Ryder U, Lamond AI. The conserved amino terminal domain of hSRP1α is essential for nuclear protein import. EMBO (Eur Mol Biol Organ) J. 1996b;15:1818–1825. [PMC free article] [PubMed] [Google Scholar]
- Wu J, Matunis MJ, Kraemer D, Blobel G, Coutavas E. Nup358, a cytoplasmically exposed nucleoporin with peptide repeats, Ran-GTP binding sites, zinc fingers, a cyclophilin A homologous domain, and a leucine-rich region. J Biol Chem. 1995;270:14209–14213. doi: 10.1074/jbc.270.23.14209. [DOI] [PubMed] [Google Scholar]
- Xiao Z, McGrew JT, Schroeder AJ, Fitzgerald-Hayes M. CSE1 and CSE2, two new genes required for accurate mitotic chromosome segregation in Saccharomyces cerevisiae. . Mol Cell Biol. 1993;13:4691–4702. doi: 10.1128/mcb.13.8.4691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama N, Hayashi N, Seki T, Panté N, Ohba T, Nishii K, Kuma K, Hayashida T, Miyata T, Aebi U, et al. A giant nucleopore protein that binds Ran/TC4. Nature (Lond) 1995;376:184–188. doi: 10.1038/376184a0. [DOI] [PubMed] [Google Scholar]