Abstract
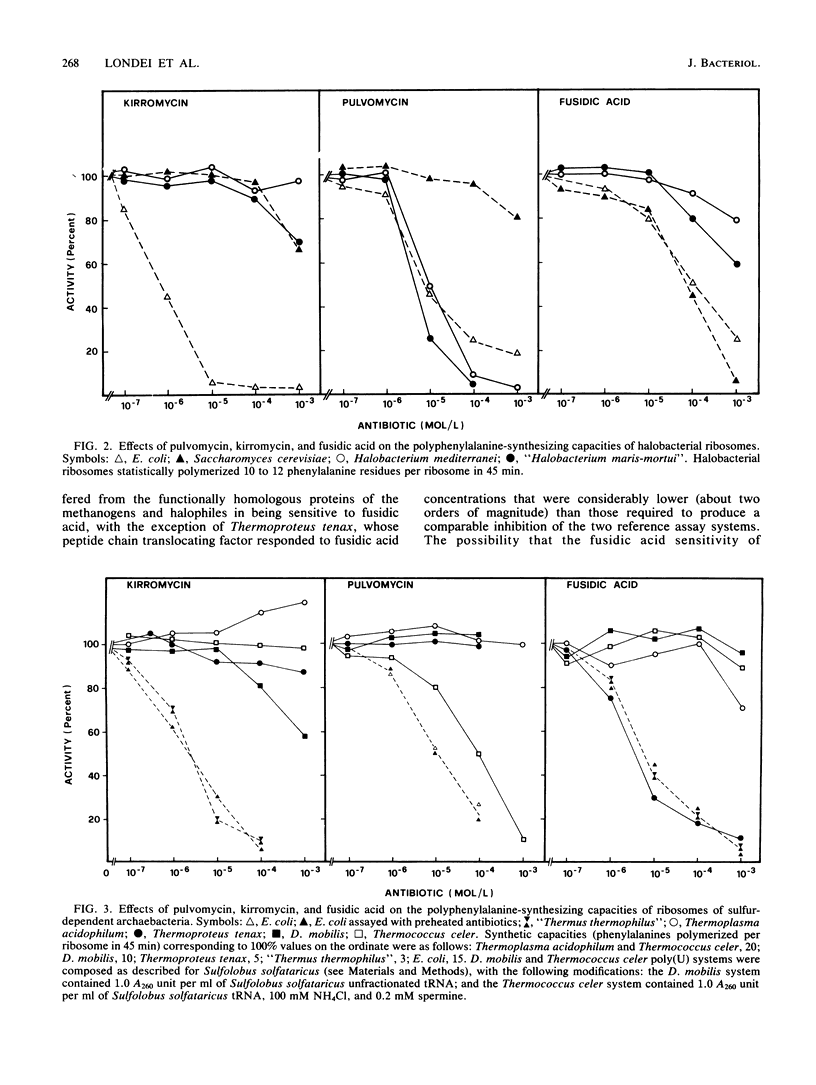
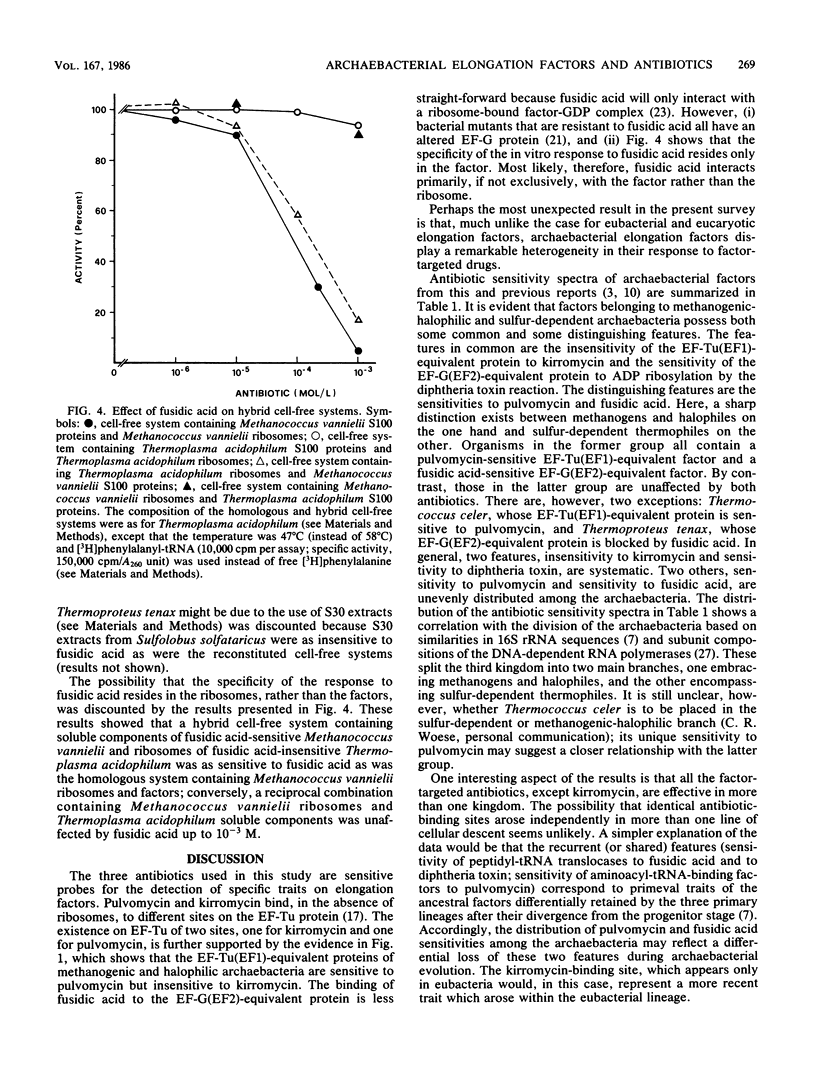
The antibiotic sensitivity of the archaebacterial factors catalyzing the binding of aminoacyl-tRNA to ribosomes (elongation factor Tu [EF-Tu] for eubacteria and elongation factor 1 [EF1] for eucaryotes) and the translocation of peptidyl-tRNA (elongation factor G [EF-G] for eubacteria and elongation factor 2 [EF2] for eucaryotes) was investigated by using two EF-Tu and EF1 [EF-Tu(EF1)]-targeted drugs, kirromycin and pulvomycin, and the EF-G and EF2 [EF-G(EF2)]-targeted drug fusidic acid. The interaction of the inhibitors with the target factors was monitored by using polyphenylalanine-synthesizing cell-free systems. A survey of methanogenic, halophilic, and sulfur-dependent archaebacteria showed that elongation factors of organisms belonging to the methanogenic-halophilic and sulfur-dependent branches of the "third kingdom" exhibit different antibiotic sensitivity spectra. Namely, the methanobacterial-halobacterial EF-Tu(EF1)-equivalent protein was found to be sensitive to pulvomycin but insensitive to kirromycin, whereas the methanobacterial-halobacterial EF-G(EF2)-equivalent protein was found to be sensitive to fusidic acid. By contrast, sulfur-dependent thermophiles were unaffected by all three antibiotics, with two exceptions; Thermococcus celer, whose EF-Tu(EF1)-equivalent factor was blocked by pulvomycin, and Thermoproteus tenax, whose EF-G(EF2)-equivalent factor was sensitive to fusidic acid. On the whole, the results revealed a remarkable intralineage heterogeneity of elongation factors not encountered within each of the two reference (eubacterial and eucaryotic) kingdoms.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bayley S. T., Griffiths E. A cell-free amino acid incorporating system from an extremely halophilic bacterium. Biochemistry. 1968 Jun;7(6):2249–2256. doi: 10.1021/bi00846a030. [DOI] [PubMed] [Google Scholar]
- Brown B. A., Bodley J. W. Primary structure at the site in beef and wheat elongation factor 2 of ADP-ribosylation by diphtheria toxin. FEBS Lett. 1979 Jul 15;103(2):253–255. doi: 10.1016/0014-5793(79)81339-3. [DOI] [PubMed] [Google Scholar]
- Cammarano P., Teichner A., Chinali G., Londei P., de Rosa M., Gambacorta A., Nicolaus B. Archaebacterial elongation factor Tu insensitive to pulvomycin and kirromycin. FEBS Lett. 1982 Nov 8;148(2):255–259. doi: 10.1016/0014-5793(82)80819-3. [DOI] [PubMed] [Google Scholar]
- Cammarano P., Teichner A., Londei P., Acca M., Nicolaus B., Sanz J. L., Amils R. Insensitivity of archaebacterial ribosomes to protein synthesis inhibitors. Evolutionary implications. EMBO J. 1985 Mar;4(3):811–816. doi: 10.1002/j.1460-2075.1985.tb03702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciferri O., Parisi B. Ribosome specificity of protein synthesis in vitro. Prog Nucleic Acid Res Mol Biol. 1970;10:121–144. doi: 10.1016/s0079-6603(08)60563-2. [DOI] [PubMed] [Google Scholar]
- Fox G. E., Stackebrandt E., Hespell R. B., Gibson J., Maniloff J., Dyer T. A., Wolfe R. S., Balch W. E., Tanner R. S., Magrum L. J. The phylogeny of prokaryotes. Science. 1980 Jul 25;209(4455):457–463. doi: 10.1126/science.6771870. [DOI] [PubMed] [Google Scholar]
- Gesteland R. F. Isolation and characterization of ribonuclease I mutants of Escherichia coli. J Mol Biol. 1966 Mar;16(1):67–84. doi: 10.1016/s0022-2836(66)80263-2. [DOI] [PubMed] [Google Scholar]
- Kessel M., Klink F. Archaebacterial elongation factor is ADP-ribosylated by diphtheria toxin. Nature. 1980 Sep 18;287(5779):250–251. doi: 10.1038/287250a0. [DOI] [PubMed] [Google Scholar]
- Kessel M., Klink F. Two elongation factors from the extremely halophilic archaebacterium Halobacterium cutirubrum. Assay systems and purification at high salt concentrations. Eur J Biochem. 1981 Mar;114(3):481–486. doi: 10.1111/j.1432-1033.1981.tb05170.x. [DOI] [PubMed] [Google Scholar]
- Klink F., Schümann H., Thomsen A. Ribosome specificity of archaebacterial elongation factor 2. Studies with hybrid polyphenylalanine synthesis systems. FEBS Lett. 1983 May 2;155(1):173–177. doi: 10.1016/0014-5793(83)80233-6. [DOI] [PubMed] [Google Scholar]
- Krisko I., Gordon J., Lipmann F. Studies on the interchangeability of one of the mammalian and bacterial supernatant factors in protein biosynthesis. J Biol Chem. 1969 Nov 25;244(22):6117–6123. [PubMed] [Google Scholar]
- NIRENBERG M. W., MATTHAEI J. H. The dependence of cell-free protein synthesis in E. coli upon naturally occurring or synthetic polyribonucleotides. Proc Natl Acad Sci U S A. 1961 Oct 15;47:1588–1602. doi: 10.1073/pnas.47.10.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pingoud A., Block W., Urbanke C., Wolf H. The antibiotics kirromycin and pulvomycin bind to different sites on the elongation factor Tu from Escherichia coli. Eur J Biochem. 1982 Apr 1;123(2):261–265. doi: 10.1111/j.1432-1033.1982.tb19762.x. [DOI] [PubMed] [Google Scholar]
- Slobin L. I. The inhibition of eucaryotic protein synthesis by procaryotic elongation factor Tu. Biochem Biophys Res Commun. 1981 Aug 31;101(4):1388–1395. doi: 10.1016/0006-291x(81)91601-6. [DOI] [PubMed] [Google Scholar]
- Swart G. W., Kraal B., Bosch L., Parmeggiani A. Allosteric changes of the guanine nucleotide site of elongation factor EF-Tu: a comparative study of two kirromycin-resistant EF-Tu species. FEBS Lett. 1982 Jun 1;142(1):101–106. doi: 10.1016/0014-5793(82)80228-7. [DOI] [PubMed] [Google Scholar]
- Tanaka N., Kinoshita T., Masukawa H. Mechanism of protein synthesis inhibition by fusidic acid and related antibiotics. Biochem Biophys Res Commun. 1968 Feb 15;30(3):278–283. doi: 10.1016/0006-291x(68)90447-6. [DOI] [PubMed] [Google Scholar]
- Tocchini-Valentini G. P., Felicetti L., Rinaldi G. M. Mutants of Escherichia coli blocked in protein synthesis: mutants with an altered G factor. Cold Spring Harb Symp Quant Biol. 1969;34:463–468. doi: 10.1101/sqb.1969.034.01.052. [DOI] [PubMed] [Google Scholar]
- Tomé F., Felicetti L., Cammarano P. Inhibition of polyphenylalanine synthesis by supernatant fractions from pea seedlings at various developmental stages. Biochim Biophys Acta. 1972 Aug 16;277(1):198–210. doi: 10.1016/0005-2787(72)90366-8. [DOI] [PubMed] [Google Scholar]
- Willie G. R., Richman N., Godtfredsen W. P., Bodley J. W. Some characteristics of and structural requirements for the interaction of 24,25-dihydrofusidic acid with ribosome - elongation factor g Complexes. Biochemistry. 1975 Apr 22;14(8):1713–1718. doi: 10.1021/bi00679a025. [DOI] [PubMed] [Google Scholar]
- Wolf H., Assmann D., Fischer E. Pulvomycin, an inhibitor of protein biosynthesis preventing ternary complex formation between elongation factor Tu, GTP, and aminoacyl-tRNA. Proc Natl Acad Sci U S A. 1978 Nov;75(11):5324–5328. doi: 10.1073/pnas.75.11.5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf H., Chinali G., Parmeggiani A. Kirromycin, an inhibitor of protein biosynthesis that acts on elongation factor Tu. Proc Natl Acad Sci U S A. 1974 Dec;71(12):4910–4914. doi: 10.1073/pnas.71.12.4910. [DOI] [PMC free article] [PubMed] [Google Scholar]


