Abstract
PTF1a is an unusual basic helix–loop–helix (bHLH) transcription factor that is required for the development of the pancreas. We show that early in pancreatic development, active PTF1a requires interaction with RBPJ, the vertebrate Suppressor of Hairless, within a stable trimeric DNA-binding complex (PTF1). Later, as acinar cell development begins, RBPJ is swapped for RBPJL, the constitutively active, pancreas-restricted paralog of RBPJ. Moreover, the Rbpjl gene is a direct target of the PTF1 complex: At the onset of acinar cell development when the Rbpjl gene is first induced, a PTF1 complex containing RBPJ is bound to the Rbpjl promoter. As development proceeds, RBPJL gradually replaces RBPJ in the PTF1 complex bound to Rbpjl and appears on the binding sites for the complex in the promoters of other acinar-specific genes, including those for the secretory digestive enzymes. A single amino acid change in PTF1a that eliminates its ability to bind RBPJ (but does not affect its binding to RBPJL) causes pancreatic development to truncate at an immature stage, without the formation of acini or islets. These results indicate that the interaction between PTF1a and RBPJ is required for the early stage of pancreatic growth, morphogenesis, and lineage fate decisions. The defects in pancreatic development phenocopy those of Ptf1a-null embryos; thus, the first critical function of PTF1a is in the context of the PTF1 complex containing RBPJ. Action within an organ-specific transcription factor is a previously unknown function for RBPJ and is independent of its role in Notch signaling.
Keywords: Ptf1a, Rbpj, Notch signaling, basic helix–loop–helix, pancreas, acinar cell
The vertebrate pancreas comprises endocrine tissue localized to the islets of Langerhans and exocrine tissues divided between acini, which make digestive enzymes, and ducts, which make a bicarbonate fluid and channel the acinar secretions to the intestine. The islets, acini, and ducts arise during embryonic development from a common pool of epithelial progenitor cells through a series of developmental transitions that are incompletely understood. In mice and rats, formation of the pancreas begins at midgestation by the evagination of mesenchyme-coated epithelial buds from dorsal and ventral positions on the posterior foregut endoderm. Most of the epithelial cells of the buds express Pdx1 and Ptf1a, key transcriptional regulators that play multiple temporally distinct roles during development and maintenance of the mature phenotype (Kim and MacDonald 2002).
The early pancreatic bud epithelium grows and branches to form a complex tubular network composed mostly of undifferentiated multipotent progenitor cells. This period of extensive growth and branching provides sufficient precursor cells for the subsequent development of the islet, acinar, and ductal tissues from this progenitor epithelium. Mutations that affect the growth and branching morphogenesis of the early epithelium prevent the formation of the islets, acini, and ducts. In embryos deficient for either PDX1 or PTF1a, the early epithelial buds grow poorly and do not mature (Jonsson et al. 1994; Offield et al. 1996; Krapp et al. 1998). For Ptf1a-null embryos, the growth of the dorsal bud epithelium is severely retarded and the ventral bud does not evaginate from the duodenum and its cells, though initially specified to become pancreas, revert to intestinal fates (Kawaguchi et al. 2002). The biochemical nature of PTF1a acting at this stage and the identity of the target genes that mediate its early effects are unknown.
During normal development, a dramatic morphogenesis termed the secondary transition (Pictet and Rutter 1972; Pictet et al. 1972) converts the simple branched tubules into a dynamic epithelium with the formation of islets segregated toward the center and acini around the periphery. The morphogenesis characteristic of the secondary transition and the initiation of the islet and acinar cell lineages require the continued presence of PDX1 (Hale et al. 2005). Precursor cells in the tubules that activate Ngn3 commit to islet fate (Gradwohl et al. 2000; Schwitzgebel et al. 2000; Gu et al. 2002), release from the tubular epithelium, and resolve into detached pre-islet cell clusters that differentiate into the five major islet cell types distinguished by the accumulation of insulin, glucagon, somatostatin, pancreatic polypeptide, or ghrelin. Pre-acini form from the ends of tubules and initiate the production of digestive hydrolases such as carboxypeptidase A1 (CPA1), amylase (AMY1), and elastase 1 (ELA1). Ptf1a expression disappears in the central pancreatic epithelium and increases in the nascent acini forming around the periphery. In mature acini, PTF1a maintains the transcription of acinar-specific genes (Krapp et al. 1996; Rose et al. 2001).
Genetic alterations that induce precocious differentiation prior to the secondary transition stop development by depleting the progenitor cell population before the branched tubule complex has a chance to form. For example, either forced expression of Ngn3, which stimulates endocrine differentiation, or genetic inactivation of components of the Notch pathway (including Rbpj), which normally antagonizes Ngn3 action, prematurely exhausts the precursor cell pool (Apelqvist et al. 1999; Jensen et al. 2000; Murtaugh et al. 2003). Conversely, persistent activation of the Notch pathway extends the undifferentiated progenitor state indefinitely and thereby prevents islet and acinar development (Hald et al. 2003).
In the mature pancreas, PTF1a (a class B basic helix–loop–helix [bHLH] protein) is present selectively in acinar cell nuclei, where it binds and activates the promoters of genes encoding the secretory digestive enzymes (Beres et al. 2006). The active form of PTF1a is part of an unusual three-subunit complex (PTF1), which also contains a common E-protein partner (such as TCF12/HEB) and RBPJL (Beres et al. 2006). RBPJL is a paralog of RBPJ, the vertebrate Suppressor of Hairless [Su(H)]. Unlike RBPJ, RBPJL is a constitutive activator that does not bind the Notch intracellular domain and therefore cannot participate in Notch signaling (Minoguchi et al. 1997; Beres et al. 2006). RBPJL is tethered to the complex through an interaction with the C-terminal domain of PTF1a (Beres et al. 2006). The trimeric PTF1 complex binds a bipartite DNA sequence composed of an E-box (CACCTG preferred) and a TC-box (TTTCCCACG), which conforms to the consensus binding sequence shared by both RBPs. The heterodimeric subcomplex of PTF1a and the E-protein binds the E-box and RBPJL binds the TC-box (Roux et al. 1989; Beres et al. 2006). Binding of the PTF1 complex to DNA is a concerted process: An isolated PTF1a:E-protein heterodimer can bind an E-box effectively, and RBPJL can bind a TC-box; however, binding by the complete trimeric complex requires both boxes.
RBPJ can form a similar trimeric complex with PTF1a that binds the same bipartite site in cell-free assays and has transcriptional activity in transfected cells (Obata et al. 2001; Beres et al. 2006); however, no function in vivo for such a complex had been found. For example, although both RBPJ and RBPJL are present in adult acinar nuclei, only the RBPJL form of PTF1 can be detected in nuclear extracts and on PTF1 target promoters (Beres et al. 2006). This unusual bHLH complex with either RBPJ or RBPJL is, so far, unique.
In addition to the roles PTF1a plays in pancreatic development, it is also necessary for the formation of the cerebellum, retina, and spinal cord (Sellick et al. 2004; Glasgow et al. 2005; Fujitani et al. 2006b). Naturally occurring mutations that truncate the C terminus of human PTF1a cause pancreatic and cerebellar agenesis (Sellick et al. 2004). The deleted portions contain overlapping domains for binding RBPJ or RBPJL, and the truncated PTF1a proteins cannot recruit either RBP into a PTF1 complex, although they still can form E-protein heterodimers capable of binding an E-box (Beres et al. 2006). These observations indicated that the failure to form a PTF1 complex disrupts the formation of the pancreas and the cerebellum, and led us to examine the biochemical form of the PTF1 complex during pancreatic development. We show here that the RBPJ form (PTF1-J) is required for the growth and morphogenesis of the dorsal and ventral pancreatic epithelia, binds and activates the Rbpjl promoter during the secondary transition, and is replaced by the RBPJL form (PTF1-L) during acinar cell differentiation. This is a previously unrecognized and Notch-independent developmental function for RBPJ.
Results
Ptf1a expression begins in the emerging epithelial buds at the onset of pancreatic development (i.e., embryonic day 9 [E9]) (Kawaguchi et al. 2002), is transiently maintained throughout the epithelium (Fig. 1A, E10.5 and E11.5; Li and Edlund 2001; Esni et al. 2004a), then disappears in the central region of the developing epithelium (E12.5), and increases in the nascent acinar crescents around the periphery (E13.5). At E14.5, Ptf1a protein and mRNA (Fig. 1B) are restricted to acini forming at the periphery of the pancreatic rudiment (Esni et al. 2004b), distinct from the interior epithelium that expresses the endocrine precursor marker Ngn3. Rbpjl mRNA is restricted to the same peripheral cell population at this developmental stage (Fig. 1B), whereas Rbpj mRNA is present throughout the epithelium (data not shown). During embryonic pancreatic development, the mRNA for PTF1a increases first, then the mRNA for RBPJL, and subsequently the mRNAs for the digestive enzymes such as ELA1 (Fig. 1C). This temporal order is consistent with the induction of Rbpjl by PTF1a, the formation of an RBPJL-containing PTF1 complex, and the subsequent induction of the acinar digestion enzymes such as ELA1.
Figure 1.
Ptf1a and Rbpjl expression during pancreatic development. (A) Immunofluorescent localization of PTF1a (green) at E10.5, E11.5, E12.5, and E13.5. (Red) E-cadherin. Arrowheads point to nascent acini. (B) Hybridization in situ detection of Ptf1a and Rbpjl mRNAs in early acinar cells around the periphery (solid arrowheads) compared with Ngn3 mRNA (open arrowheads) in endocrine cell precursors in the interior of the pancreatic epithelium at E14.5. Ptf1a and Rbpjl are for nearby, nonadjacent sections. (C) mRNAs were quantified by quantitative RT–PCR with RNA isolated from embryonic pancreatic rudiments and expressed relative to the levels at E18.5. The Rbpj and Rbpjl level mRNAs have a reciprocal relationship. Error bars are SEM; points without bars have SEMs equal to or smaller than the symbol.
The RBPJL form of the PTF1 complex binds and activates the Rbpjl promoter
To determine whether PTF1a might indeed control the transcription of Rbpjl, we searched the 10-Kb 5′ flanking region and intronic sequences of the Rbpjl gene for potential PTF1-binding sites comprising an E-box and a TC-box spaced one or two helical DNA turns apart (Cockell et al. 1989; Rose and MacDonald 1997; Beres et al. 2006). Two potential binding sites with sequences that conform to the consensus were detected; both are within 1.4 Kb of the transcriptional start site (Fig. 2A). The proximal PTF1-binding site near the transcriptional start (at −91) is composed of an RBP-binding sequence with tandem E-boxes one and two helical turns away, and is retained from birds to primates. The distal site (at −1341) has an E-box two DNA turns from an RBP-binding sequence and is conserved well only among mammals.
Figure 2.
Properties of the two PTF1-binding sites centered at −1341 and −92 of the mouse Rbpjl gene. (A) The nucleotide sequence of the two sites in the Rbpjl promoter are conserved. The distal site has an E-box separated from a TC-box by 21–22 bp, center-to-center; the proximal site has tandem E-boxes separated by 11 and 22 bp from a single TC-box. (B) Both Rbpjl promoter sites can bind PTF1 in nuclear extracts from adult pancreas. Binding is compared with that with the well-characterized PTF1 site from the Ela1 enhancer. Supershift analyses showed that all three sites tested bind the complete PTF1 complex (a trimer comprising PTF1a, a common E-protein [predominantly TCF12] and RBPJL) as well as a partial complex (a heterodimer of PTF1a and TCF12). (C) Comparison of the activities of the Ela1 and Rbpjl PTF1-binding sites in transfected cells. A tandem repeat of the 26-bp Ela1 or the 32-bp proximal Rbpjl PTF1 site was linked to the minimal promoter (−92 to +8) from the Ela1 gene driving a luciferase reporter gene and transfected into either the mouse 266-6 pancreatic acinar cell line or the human 293 embryonic kidney cell line. The activity of each construct was corrected for transfection efficiency and calculated relative to the activity of the same reporter, but lacking a PTF1-repeat. Error bars are standard deviations. (D) A model for the two PTF1 complexes and DNA binding that distinguishes the overlapping regions of PTF1a that interact with RBPJ or RBPJL and the basis for the differential effects of the W298A mutation. The Ws indicate the two tryptophan-containing conserved peptide motifs in the C-terminal region of PTF1a that are primary determinants for binding the RBPs. The W298A mutation is indicated by the A; based on the results of Beres et al. (2006).
Both PTF1 sites can bind the authentic RBPJL form of the PTF1 complex isolated from adult pancreas, and the complex can be supershifted by antibody against PTF1a (Fig. 2B). Moreover, the two sites also bind PTF1 complexes reconstituted from the individual subunits synthesized by cell-free translation. Binding requires both an E-box and the TC-box with proper spacing (see Fig. 6, below; data not shown).
Figure 6.
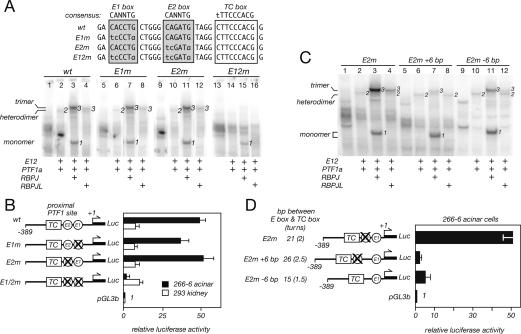
The two E-boxes of the proximal PTF1-binding site are redundant. (A) Mutational disruption of the first (E1m) or second (E2m) E-box had no effect on the binding of the PTF1a/E12 heterodimer (cf. lanes 2,6,10), the RBPJ form of the trimer (lanes 3,7,11), or the RBPJL form of the trimer (lanes 4,8,12). (Lanes 14–16) Mutation of both E-boxes (E12m) eliminated binding of the dimer and both forms of the trimer. The positions of the monomer RBPJ (1), the PTF1a/E12 heterodimer (2), and the trimers (3) are indicated. The PTF1 subunits were synthesized by cell-free translation. (B) The E-box mutations shown in A were incorporated into the −389 RbpL-Luc plasmid to test whether both E-boxes were also required for activity of the Rbpjl promoter in transfected acinar or kidney cells. Mutational inactivation of either E-box had no effect on acinar activity, whereas inactivation of both nearly eliminated it. None of the mutations affected activity of the promoter in 293 kidney cells. (C) A proper stereochemical relationship between the TC-box and an E-box is required for binding the complete PTF1 trimer in EMSAs. To simplify the complexity of two E-boxes, we mutated the proximal one (which had no discernible effect on complex binding or activity) and tested the effects of altered spacing between the TC-box and the remaining E-box by inserting 6 bp (E2m + 6 bp) or deleting 6 bp (E2m − 6 bp) (see the diagram in D). Either alteration decreased binding of the PTF1 trimer, but not the PTF1a/E12 heterodimer. (D) Proper spacing between the TC-box and an E-box is required for the acinar activity of the promoter. The spacing changes described for C were incorporated into −389 RbpL-Luc for transfection into 266-6 acinar cells. Either alteration decreased acinar activity by at least 90%. Error bars are standard deviations.
A 1.4-Kb Rbpjl promoter region containing the transcriptional start and both potential PTF1-binding sites increased the activity of the promoterless reporter plasmid pGL3b at least 140-fold in two acinar cell lines tested by transfection (Fig. 3A,B). Mutational inactivation of the TC-box (Fig. 3B) or the E-boxes (Fig. 3C) of the proximal site eliminated the acinar activity, whereas similar mutations of the distal site had no affect. The mutations had only modest effects on the low activity of the 1.4-Kb promoter in the nonpancreatic 293 kidney cell line. A 367-base-pair (bp) promoter region that retains the proximal but not the distal site also retained high acinar-specific activity (Fig. 3A). Therefore, in this assay, the proximal PTF1-binding site is critical for activity of the promoter in acinar cells, but appears to be irrelevant in nonacinar cells. It is noteworthy that this is the first PTF1-binding site recognized that has two E-boxes, which we later show are redundant (see Fig. 6, below).
Figure 3.
Activity of the Rbpjl promoter in acinar cell lines. (A) Acinar cell activity is retained within the proximal 389 bp. The promoter regions extend from the distal points shown to 103 nucleotides of the 5′ untranslated region of Rbpjl and linked to the reporter vector pGL3-basic for transfection into acinar and nonpancreatic cell lines. The position of the first exon of the nearby Matrilin4 gene, transcribed in the direction opposite to Rbpjl, is shown. (B) Inactivation of the proximal PTF1-binding site by mutation of the TC-box eliminates the acinar activity; mutation of the distal PTF1 site had no discernible effect. The effects were similar for two independently derived pancreatic acinar tumor cell lines, mouse 266-6 and rat AR4-2J. The mutations, indicated by Xs, were introduced in the context of the −1380-bp promoter (hereafter, 1.4K). (C) Inactivation of the proximal PTF1-binding site by mutation of the two E-boxes eliminates the acinar activity; mutation of the distal site had no effect. For the experiments shown in A–C, the promoter alterations tested had no significant effect in the nonpancreatic 293 cell line. Luciferase reporter activity was adjusted for transfection efficiency and expressed relative to the promoterless pGL3-Basic vector. Error bars are standard deviations.
Not only was the proximal PTF1-binding site necessary to drive acinar cell transcription, but it was also sufficient when multimerized and linked to a passive minimal promoter (Fig. 2C). The acinar activity for the multimerized PTF1-binding site from the Rbpjl promoter could be reconstituted in 293 cells by the ectopic expression of the PTF1 subunits (see Fig. 7A, below). Thus, the proximal PTF1-binding site is an acinar-specific promoter element that is highly responsive to components of the PTF1 complex.
Figure 7.
The activity of the proximal PTF1 site can be reconstituted by ectopic expression of the PTF1 subunits. (A) Coexpression of PTF1a, HEB, and RBPJ or RBPJL in 293 kidney cells induces transcription from a synthetic promoter containing three copies of the proximal PTF1 site from the Rbpjl promoter (described in Fig. 2C). The activity in response to PTF1a alone was due to the presence of substantial RBPJ and E-proteins in 293 kidney cells. The activity was enhanced by the overexpression of HEB/TCF12 or RBPJL (which is absent in 293 cells). (B) The activities of the PTF1 sites from the Rbpjl and Ela1 promoters differ quantitatively and qualitatively. The Rbpjl site is many-fold more active than the Ela1 site in response to either the J or the JL forms of the reconstituted PTF1 (note the different scales). Moreover, the difference in the ability of the J and JL forms to activate the Ela1 site (approximately sixfold) is nearly absent for the Rbpjl site (approximately twofold). Error bars are standard deviations.
To determine whether either PTF1-binding site is active in vivo, we tested the normal and mutated 1.4-Kb regions in founder transgenic embryos (Fig. 4). Of the 22 transgenic embryos obtained from fertilized eggs microinjected with the normal transgene, eight (36%) stained intensely for the transgenic β-galactosidase reporter in the pancreas, but nowhere else in the viscera. A mutation of the proximal site sufficient to eliminate PTF1 binding abolished expression of the transgene, whereas the equivalent mutation of the distal PTF1 site had little if any effect (25% expressed). Thus, the proximal site is also critical for expression in the pancreas of mouse embryos, and the relative importance of the two sites was paralleled in vivo and in vitro.
Figure 4.
Activity of the Rbpjl promoter in vivo. Transgenic embryos at 17.5 d of gestation were derived from implanted, fertilized eggs injected with reporter constructs containing the 1.4-Kb promoter without alterations (A) or with the mutations described in Figure 3B of the TC-box of the proximal (B) or distal (C) PTF1-binding site. The expression of each transgene was detected by β-galactosidase activity derived from the lacZ reporter gene. The duodenum (D) has a low level of endogenous β-galactosidase activity; to verify the absence of pancreatic expression from the transgene with the mutated proximal PTF1 site, the staining was extended for B. (dP) Pancreas derived from the dorsal bud; (vP) pancreas from the ventral bud; (St) stomach; (Sp) spleen; (D) duodenum. (Bottom) The number of embryos with pancreatic β-galactosidase activity out of the total number of transgenic embryos is a measure of promoter potency.
The PTF1 complex can be formed with either RBPJ or RBPJL (see Fig. 2D; Beres et al. 2006). In the adult pancreas, the RBPJL form of the complex is bound to the promoters of the digestive enzyme genes, including Ela1 (Beres et al. 2006). The results from immunoprecipitation of chromatin from adult mouse pancreas demonstrated that both PTF1a and RBPJL were present on the proximal Rbpjl promoter region containing the PTF1-binding site, whereas RBPJ was not (Fig. 5A). Consecutive immunoprecipitations of pancreatic chromatin, first with the antibody for PTF1a, and then the antibody for RBPJL (and vice versa), enriched the Rbpjl promoter equivalent to the arithmetical product of the individual enrichments (Fig. 5B), which is consistent with coresidence of the two proteins at the same allele (Geisberg and Struhl 2004). These results indicate that RBPJL acts within the PTF1 complex to autoactivate the transcription of its gene in adult acinar cells.
Figure 5.
PTF1 subunits reside on the Rbpjl proximal promoter region containing the PTF1 site in chromatin from adult pancreas. (A) ChIP analysis using antibodies to individual subunits showed that PTF1a and RBPJL, but not RBPJ, are present at or near the proximal PTF1 site. Results from a similar experiment showed PTF1a and RBPJL present at the distal PTF1 site as well (data not shown). Binding could not be detected using chromatin from adult liver. The schematic of the 1.4-Kb Rbpjl promoter shows the relative positions of the PTF1-binding sites and the primer pairs for PCR amplification. (B) Sequential ChIPs indicated that PTF1a and RBPJL coreside on Rbpjl promoters in adult acinar cells. The specificity of the consecutive enrichments was verified by the lack of enrichment with consecutive precipitations using an antibody to an unrelated DNA-binding protein (i.e., cMYC) and by the loss of any initial purification with anti-PTF1a or anti-RBPJL when anti-cMyc was used as the second precipitating antibody. Fold enrichment was quantified by quantitative PCR; a zero indicates that promoter DNA was not detected and a one indicates no enrichment relative to the input DNA.
The unusual nature of the proximal PTF1-binding site
The trimeric PTF1 complex forms in solution and subsequent binding to DNA requires an E-box and a TC-box separated by one or two helical turns of DNA (Beres et al. 2006). These properties are diagnostic of the concerted binding of the PTF1 complex and distinguish the binding of an intact complex from the independent binding of individual components or subcomplexes. The pairing of two E-boxes with a TC-box in the proximal site was unprecedented among known PTF1-binding sites, and we tested whether both E-boxes are necessary for complex binding and transcriptional activation. The results of EMSA analyses showed that either E-box was effective in combination with the TC-box, because disruption of both E-boxes was required to abolish binding of a reconstituted trimeric PTF1 complex containing either RBPJ or RBPJL (Fig. 6A, cf. lanes 3,7,11 and 15, and cf. lanes 4,8,12 and 16). Similarly, the disruption of both E-boxes was required to eliminate the activity of the −389 proximal promoter fragment in transfected acinar cells (Fig. 6B).
Mutation of both E-boxes also eliminated the binding of the PTF1a:E12 heterodimer (Fig. 6A, cf. lanes 2,6,10 and 14). To confirm that the critical requirement for an E-box is the binding of a trimeric PTF1 complex and not only the PTF1a–E12 heterodimer, we tested the effects of changing the spacing between an E-box and a TC-box. Inserting or removing a half DNA helical turn reduced binding of both the J and JL forms of the trimeric PTF1 (Fig. 6C) without affecting the independent binding of the PTF1a:E12 dimer (Fig. 6C, cf. lanes 2,6,10) or RBPJ (Fig. 6C, cf. the monomer band in lanes 3,7,11). The half-turn changes also decreased the activity of the Rbpjl promoter 90%–97% in transfected acinar cells (Fig. 6D). These results verify that the activity of the PTF1-binding site in the Rbpjl promoter requires the concerted binding of a trimeric complex.
To examine the regulatory properties of the isolated proximal PTF1-binding site, we compared the activity of a three-copy repeat of the site with that of three copies of the well-characterized PTF1 site from the Ela1 promoter (Rose and MacDonald 1997; Rose et al. 2001; Beres et al. 2006). Each repeat was linked to the minimal promoter of the Ela1 gene driving a luciferase reporter and assayed by transfection into the 266-6 acinar cell line or the 293 kidney cell line. The PTF1 site from the Rbpjl promoter was highly active in 266-6 acinar cells, compared with the site from the Ela1 promoter (see Fig. 2C). The acinar activity of the Rbpjl site could be reconstituted in 293 kidney cells by the ectopic expression of the PTF1 subunits (Fig. 7A). Although the Rbpjl site was much more active than the Ela1 site with either PTF1 form, its greater effectiveness with the J form (90-fold) was particularly striking compared with the Ela1 site (sixfold) (Fig. 7B). The Rbpjl version is favored because its TC-box is identical to the optimal RBP-binding sequence (Supplementary Fig. S1). We reasoned that the enhanced ability to be activated by the J form of the PTF1 complex might be important for the acinar-specific induction of Rbpjl during pancreatic development.
PTF1 switches from an RBPJ to an RBPJL form during pancreatic development
To examine whether a PTF1 complex containing RBPJ may be the initial activator of Rbpjl transcription, we monitored the presence of PTF1a, RBPJL, and RBPJ on the proximal Rbpjl promoter just prior to (E12.5), during (E15.5), and after (E17.5) the secondary transition (Fig. 8A). PTF1a and RBPJ were present early at E12.5, when acinar cell development is beginning and Rbpjl is activated. As development progressed, the ratio of RBPJL to RBPJ on the promoter increased seven- to eightfold. Thus, the PTF1 complex bound to the Rbpjl promoter changes from one containing RBPJ, when the promoter is being activated, to one with RBPJL, as the RBPJL protein accumulates. This switch does not occur for all PTF1 targets, however; only the RBPJL form of PTF1 is detected on the CPA1 promoter in the developing pancreas and is retained in adult acinar cells (Fig. 8B). It appears that the production of RBPJL is an early event in acinar development that then contributes to the activation of the digestive enzyme genes.
Figure 8.
RBPJ is exchanged for RBPJL on the Rbpjl promoter during pancreatic development. ChIP analysis of PTF1a, RBPJL, and RBPJ at the proximal promoter region of Rbpjl (A) and the proximal Cpa1 promoter (B). An antibody to rabbit IgG was used to estimate background precipitation of Rbpjl promoter DNA. Arrows flanking the PTF1-binding sites in the promoters indicate the positions of the PCR primers. Error bars are SEM for three ChIPs.
The early developmental function of PTF1a requires its interaction with RBPJ
Previously (Beres et al. 2006), we showed that the substitution of alanine for tryptophan at residue 298 in the PTF1a-binding site for RBPJ extinguished the ability of PTF1a to interact with RBPJ, yet retained its ability to interact with RBPJL (see Fig. 2D). This PTF1a mutant provides the means to investigate the role of the J form of the trimeric PTF1 complex independent of the JL form. To determine whether the J form of the PTF1 complex is required for the early growth and branching morphogenesis phase of pancreatic development, we generated a line of mice bearing the W298A mutation (Ptf1aWA) using recombination-mediated cassette exchange (Long et al. 2004; J. Burlison, Q. Long, C.V.E. Wright, and M.A. Magnuson, in prep.). As for mice heterozygous for the Ptf1acre-null allele (Kawaguchi et al. 2002), pancreatic development and function for heterozygous Ptf1aWA/+ mice were normal. However, the pancreatic development of embryos that expressed only the W298A form of PTF1a was severely affected and mirrored that of homozygous Ptf1a-null embryos (see below).
We used the activation of the Rosa26R locus and β-galactosidase staining of Ptf1aWA/cre embryos to follow the fate of the PTF1a-deficient cells initially specified for pancreas and the growth of the mutant epithelium (Fig. 9). The cre allele has the entire coding region of Ptf1a replaced by that of cre, and therefore is null for PTF1a function (Kawaguchi et al. 2002). The developmental defects of Ptf1aWA/cre embryos were indistinguishable from those of Ptf1aWA/WA embryos (Supplementary Fig. S2). At E10.5, the dorsal bud of the Ptf1aWA/WA pancreas appeared normal, including the size of the epithelium and the presence of PDX1 (see Fig. 10, top). A Ptf1a expression domain was present in the duodenal epithelium at the normal position of the ventral bud, but budding did not occur (data not shown). Indeed, as for Ptf1acre/cre-null embryos (Kawaguchi et al. 2002), the ventral pancreas never formed. Rather, pancreatic precursor cells that activated the Ptf1a locus became marked by β-galactosidase activity and reverted to an intestinal fate within the duodenal epithelium (Fig. 9C′, inset; data not shown). Developmental defects of the dorsal epithelial bud in Ptf1aWA/cre embryos became evident at E11.5. As for Ptf1acre/cre embryos (Fig. 9A), the growth of the dorsal epithelium was retarded (Fig. 9A′); by E12.5, the epithelium was markedly smaller than normal (Fig. 9B′). Because the PTF1aWA protein was present (see Fig. 10B′), developmental defects were not due to an inability to accumulate the mutant protein. At 1 d prior to birth, the dorsal pancreatic epithelium was a truncated, poorly branched, duct-like structure (Fig. 9C′) with columnar epithelial cells, akin to the ductal vestiges of Ptf1a-deficient embryos (Krapp et al. 1998; Kawaguchi et al. 2002). Because the W298A mutant so closely phenocopies the null, it appears that the first crucial function of PTF1a during pancreatic development requires its interaction with RBPJ.
Figure 9.
Defective pancreatic development for the W298A mutant of Ptf1a. Pancreatic tissue derived from Ptf1a-expressing endoderm is stained for β-galactosidase activity induced by Ptf1a-cre activation of the R26R locus. (A–C) Normal pancreatic development of heterozygous Ptf1a+/cre embryos at E11.5, E12.5, and E17.5. (C) The normal perinatal pancreas is composed of two lobes derived from the dorsal pancreatic bud (dP) and one from the ventral pancreatic bud (vP), which lies within the first loop of the duodenum (D). A includes a view of the truncated pancreatic development of a homozygous Ptf1acre/cre embryo for comparison. (A′–C′) Abnormal pancreatic development of Ptf1aWA/cre embryos. One Ptf1acre allele is necessary to follow the fate of the mutant pancreatic cells; homozygous Ptf1aWA/WA embryos have an identical mutant phenotype (Supplementary Fig. S2). (A′) Altered pancreatic development is evident at E11.5, and is indistinguishable from that of homozygous-deficient Ptf1acre/cre embryos. (B′) Compared with normal development at E12.5, growth of the dorsal pancreatic epithelium is curtailed, and a ventral epithelium has not escaped the duodenal mucosa. (C′) The dorsal pancreas forms a truncated, undeveloped ductal structure (dP) without islet or acinar tissues. A ventral pancreas does not form; rather, the mutant, β-galactosidase-positive cells incorporate into the gut mucosa and acquire intestinal phenotypes (vP). (Inset) Cells within the normal duodenal epithelium stain for β-galactosidase activity and occasionally costain red with PAS, indicative of goblet cells. (dP) Dorsal pancreas; (vP) ventral pancreas; (D) duodenum; (St) stomach; (Sp) spleen.
Figure 10.
Loss of pancreatic development markers in homozygous W298A mutant embryos. (Top) For Ptf1aWA/WA embryos at E10.5 (A′–D′), epithelial morphogenesis (A,A′) of the dorsal pancreas and the levels of PTF1a (B,B′) and PDX1 (C,C′) were indistinguishable from those for normal development of heterozygous Ptf1a+/WA (A–D) and wild-type embryos (data not shown). By E14.5 in mutant embryos, morphogenesis was defective (E,E′), PTF1a did not appear in newly forming acini (F,F′,H,H′; acini), and cells with high levels of PDX1 were absent (G,G′). (A,A′,E,E′) E-cadherin delineates the pancreatic epithelium. (Bottom) Neither islets nor acini form for Ptf1aWA/WA embryos. At E14.5, CPA1, an early marker of acinar cell differentiation (I; arrow), was absent from mutant embryonic pancreas (I′). Endocrine cell clusters in the mutant pancreas coexpress prohormone convertase 1/3 (PC1/3) (e.g., arrow, J′) and glucagon (arrow, K′). Insulin-expressing islet β-cells are normally present at high numbers (inset of L; insulin, blue), but are rare in the mutant (inset of L′; insulin, blue). Bars, 50 μm.
The inability of PTF1a to bind RBPJ prevents the secondary transition
Normal growth and morphogenesis of the early pancreatic epithelia establish an extensive precursor cell population that fuels the genesis of islet and acinar cells during the secondary transition. Exhausting this precursor population by forcing precocious differentiation (Apelqvist et al. 1999; Hald et al. 2003; Murtaugh et al. 2003) or preventing its formation by inactivation of Pdx1 (Ahlgren et al. 1996; Hale et al. 2005) or Ptf1a (Krapp et al. 1998; Kawaguchi et al. 2002) precludes this developmental transition. Because the growth and branching of the early Ptf1aWA/WA pancreas are also greatly attenuated, we examined developmental markers to determine whether the secondary transition occurs (Fig. 10). The appearance of PTF1a in proacini emerging during the secondary transition (Fig. 10F) did not occur in the mutant embryos (Fig. 10F′). Pdx1 and Sox9 expression continued in the mutant (Fig. 10G′; data not shown), although at the low levels characteristic of the precursor epithelium prior to the secondary transition. Acinar structures did not form, and acinar differentiation markers such as CPA1 did not appear (Fig. 10I′). The characteristic surge of endocrine cells also did not occur. The induction of high PDX1 levels that normally occurs in precursor cells directed to the β-cell lineage (Fujitani et al. 2006a) did not occur in the mutant and the insulin-producing islet β-cells that normally form in large numbers at this stage are far fewer in the mutant (see the insets of Fig. 10L, L′). The endocrine cell clusters present in the mutant pancreas appeared early, prior to the secondary transition, and were composed of glucagon+ cells that also contained prohormone convertase 1/3 (PC1/3) (Fig. 10J–L, J′–L′) rather than PC2. The early appearance and the combination of markers indicate that these cell clusters are of the early endocrine lineage (Kim and MacDonald 2002; Wilson et al. 2002) rather than the islet lineage. Thus, neither islet nor acinar developmental programs begin in the mutant pancreas.
Discussion
In this report we showed that Ptf1a controls pancreatic development at three distinct phases in two distinct biochemical forms. The PTF1 complex containing RBPJ (PTF1-J) is required first for the growth and branching morphogenesis of the epithelium of the nascent pancreatic buds. This is a previously unknown developmental function for RBPJ, in which it does not act as the transcriptional mediator of Notch signaling. At approximately E13, when Ptf1a expression is enhanced in the newly forming acinar cell clusters, RBPJL appears and displaces RBPJ in active transcription complexes. This coincides with, or slightly precedes, the onset of expression of the digestive hydrolase genes, which characterize the differentiated acinar cells. It is the RBPJL form of the PTF1 complex (PTF1-L) that is bound to the active CPA1 promoter in the developing pancreas and in mature acinar cells. Thus, during normal pancreatic development, it appears that a switch from PTF1-J to PTF1-L mediates the activation of other acinar-specific genes. In the mature gland, PTF1-L and not PTF1-J is detected on all of the digestive enzyme genes examined.
Notch-dependent and Notch-independent functions of RBPJ
Compelling evidence for Notch signaling through RBPJ during pancreatic development derives from the effects of genetic alterations of Notch pathway components. Germline mutations that inactivate genes encoding components of the canonical Notch pathway (i.e., Dll1, Hes1, Rbpj, or Notch1) disrupt early pancreatic growth and branching, deplete epithelial precursor cells by overcommitment to the early endocrine lineage, and eliminate or delay the secondary transition and consequently the formation of islets and acini as well (Apelqvist et al. 1999; Jensen et al. 2000; Fujikura et al. 2006). Similar developmental defects occur by the premature overexpression of the proendocrine Ngn3 gene, which overrides Notch control by antagonizing Hes1 (Jensen et al. 2000; Schwitzgebel et al. 2000; Lee et al. 2001). Conversely, misexpression of activated Notch locks the progenitor cells of the pancreatic epithelium into an undifferentiated state (Hald et al. 2003; Murtaugh et al. 2003). Thus, Notch signaling is required for proper pancreatic development, and RBPJ is the transcriptional mediator. However, these genetic experiments do not distinguish Notch-dependent from Notch-independent roles for pathway components such as RBPJ.
Several lines of evidence had suggested that such a role for RBPJ was likely. (1) Using RBPJ as bait, Obata et al. (2001) caught PTF1a in a yeast two-hybrid screen with a cDNA library from embryonic neural tissue and showed that coexpression of PTF1a and RBPJ could activate a reporter gene driven by the PTF1-binding site from the Ctrb promoter in transfected cells. (2) RBPJ can form a trimeric complex with PTF1a and an E-protein that can bind a known PTF1-L target site and activate transcription through that site in transfected cells (Beres et al. 2006). (3) C-terminal truncations of the human PTF1a that eliminate binding of either RBP (Beres et al. 2006) disrupt the development of the cerebellum (Sellick et al. 2004) where RBPJ is present but RBPJL is not. (4) Orthologs of PTF1a, E2A, and RBPJ (but not RBPJL) are conserved to insects and are able to form an equivalent trimeric complex with identical DNA-binding properties and transactivation potential in mammalian cells (Beres et al. 2006). Thus, PTF1-J appears to be the ancestral complex.
Because PTF1a is required for pancreatic development prior to the appearance of RBPJL, we reasoned that RBPJ might have a Notch-independent function as an obligate partner for PTF1a early, and that the Rbpjl gene might be a target of the complex. We found two potential PTF1-binding sites in the Rbpjl promoter. The proximal site could bind and be activated by either the PTF1-J or PTF1-L, and mutations in this site disrupted promoter activity in transfected acinar cells and the pancreas of transgenic embryos. Moreover, PTF1-J was present on the Rbpjl promoter at the onset of acinar cell differentiation when transcriptional activation of Rbpjl begins, and was gradually replaced by the JL form as differentiation progressed and mature acinar cells accumulated.
To test the importance of the J form of PTF1 directly, we derived a line of mice that express a mutant PTF1a with a single amino acid change (W298A) that disrupts its interaction with RBPJ, but not RBPJL. Pancreatic development of homozygous Ptf1aWA/WA embryos was arrested in the same manner as Ptf1a-null embryos, indicating that the first critical function of PTF1a requires the partnership of RBPJ within PTF1-J, a role for RBPJ distinct from that in Notch signaling.
Although effects on pancreatic development of Rbpj−/− and Ptf1aWA/WA embryos are similar, important differences indicate that RBPJ is required as a partner for Notch as well as for PTF1a, with roles that overlap temporally but not functionally. In the absence of RBPJ, pancreatic epithelial growth is slowed by the premature and enhanced formation of differentiated endocrine cells (Apelqvist et al. 1999; Fujikura et al. 2006) due to the unrestricted differentiation of the early endocrine lineage. The Rbpj−/− defects are identical to those of null mutations for the other Notch pathway components Delta-like1 and Hes1 (Apelqvist et al. 1999; Jensen et al. 2000) and are consistent with the expected consequence of lost Notch signaling, which is premature differentiation due to an inability to sustain a population of progenitor cells. In mutant embryos with the version of PTF1a that is unable to bind RBPJ, epithelial growth and branching are also attenuated, and the early endocrine cells are present. These similar developmental abnormalities indicate that the principle cause of the pancreatic defects for the W298A version of PTF1a is its inability to interact with RBPJ, although a formal possibility remains that an interaction of PTF1a with an as yet unidentified partner is also disrupted. Because Notch signaling remains intact for Ptf1aWA/WA embryos, the early pancreatic endocrine population is not expanded as it is in the Rbpj-deficient embryos. This important difference indicates that RBPJ serves two separate functions in pancreatic development, one for Notch control of the progenitor state and another for PTF1 control of the early developmental program. Rbpj mutants are deficient for both, whereas the inability to recruit RBPJ into the PTF1 complex only affects the latter.
The tryptophan to alanine mutation is near the C terminus in a heptapeptide sequence that is conserved between vertebrates and flies. The PTF1a motif (WTPEDPR) is similar to the critical sequence in the NotchICD (WTPEGFK) for binding to RBPJ (Kovall and Hendrickson 2004). Indeed, a synthetic peptide containing the Notch motif inhibits the recruitment of RBPJ by PTF1a, and overexpression of either the NotchICD or PTF1a in cell transfection experiments interferes with the activity of the other (Beres et al. 2006). It would be instructive to know whether this mutual antagonism has a regulatory function during development. The transition to RBPJL in acini may serve to unlink the RBP subunit from Notch signaling. Alternatively, the selective use of the RBPJL in the PTF1 complex of adult acini may ensure the availability of RBPJ for its Notch pathway function.
The developmental abnormalities caused by the W298A form of PTF1a prove our model (Beres et al. 2006) for the defects in human PTF1a shown to be associated with permanent neonatal diabetes mellitus (PNDM) (Sellick et al. 2004). Two human mutations delete short regions of the C terminus of PTF1a containing an evolutionarily conserved hexapeptide sequence that includes W298. Neither mutant protein is able to recruit RBPJ into a trimeric PTF1 complex (Beres et al. 2006). Our results show that a much subtler mutation with the same biochemical consequences causes the same defects of pancreatic and cerebellar development as the PNDM newborns (this study; H. Kei, Y. Nakada, S.M. Glasgow, T. Masui, M. Henke, T.M. Beres, M.A. Magnuson, R.J. MacDonald, and J.E. Johnson, in prep.).
Autoregulation of Rbpjl
The proximal PTF1-binding element of the Rbpjl promoter is required for the activation of Rbpjl transcription in nascent acinar cells. The presence of PTF1a and RBPJ on the proximal promoter region at this time indicates that PTF1-J initiates this first stage of Rbpjl expression. The presence of RBPJ in an organ-specific transcription factor complex is novel. As acinar differentiation progresses, RBPJL gradually replaces RBPJ on the promoter. Because transcription is begun by PTF1-J and continued by PTF1-L, the switch must occur in the same cell, either by an exchange of RBP subunits or of complete PTF1 complexes.
Several lines of experimental evidence indicate that a complete trimeric PTF1 complex binds and activates the Rbpjl promoter rather than individual subunits that bind and act independently. PTF1a and RBPJL coreside within the Rbpjl promoter region containing a consensus binding site for the trimeric PTF1 complex essential for the acinar activity of the promoter; the vast majority of the RBPJL DNA-binding activity in nuclear extracts from adult acinar cells is in PTF1-L (Beres et al. 2006); the formation of a stable trimeric complex with highly cooperative binding properties occurs prior to binding to DNA; alterations of the spacing between the E-box and TC-box that prevent binding of either PTF1 complex, but allow independent binding of a PTF1a–HEB heterodimer to the E-box and RBPJL (or RBPJ) to the TC-box, eliminate the transcriptional activity of the PTF1-binding site. The sum of this evidence indicates that the trimeric PTF1 complex mediates the cell-specific activity of the Rbpjl promoter in mature acinar cells.
The autoactivation of Rbpjl by PTF1-L may provide a stable autoregulatory loop that ensures the maintenance of the acinar cell phenotype. PTF1-L is also present on PTF1-binding sites necessary for acinar transcription of all the pancreatic digestive enzyme genes examined (Beres et al. 2006). Thus, the stable maintenance of Rbpjl expression can directly control the continued transcription of genes that define the acinar cell phenotype. Preliminary observations of a Rbpjl knockout line of mice (T. Masui, M.E. Magnuson, and R.J. MacDonald, unpubl.) indicate that the mutant pancreas at birth is consistently one-third less than normal and acinar tissue is less differentiated, as judged by 70% lower levels of the mRNAs for two of the five digestive enzymes so far examined. Thus, RBPJL is important for a maximal acinar phenotype, though not critical for formation of acinar cells. It will be interesting to determine whether RBPJL is required for an optimal response to dietary changes or for acinar regeneration after injury.
The PTF1 complex has an ancient function
An orthologous trimeric PTF1 complex is conserved to insects (Beres et al. 2006). Drosophila melanogaster has a PTF1a ortholog (FER1) that can a form a complex with the Drosophila E2A equivalent (Daughterless; DA) and Su(H). The Drosophila complex can bind the PTF1 site from the mouse Ela1 promoter and has transactivation potential in transfected mammalian cells intermediate to the mammalian PTF1-J and PTF1-L. Remarkably, the conservation is so great that mixed complexes of Drosophila and mammalian subunits are functional.
Insects do not have a digestive gland homologous to the vertebrate pancreas. The pancreatic endocrine function is served by cells in the brain that deliver insulin-like peptides to the heart tube and gut and neuroendocrine cells of the ring gland that release peptides with glucagon-like functions into the hemolymph (Rulifson et al. 2002; Kim and Rulifson 2004). Although digestive serine proteases are produced in the ventriculus and gastric caecae (Davis et al. 1985), Fer1 is not expressed there (http://flybase.bio.indiana.edu; J. Zoloty, R.J. MacDonald, and D.M. McKearin, unpubl.), and most digestive hydrolases are made in the midgut mucosa. Consequently, Fer1 does not have a regulatory role equivalent to that of Ptf1a in pancreatic development and acinar function. Rather, the extensive expression of both Fer1 (http://flybase.bio.indiana.edu) and Ptf1a (Obata et al. 2001; Sellick et al. 2004; Glasgow et al. 2005) in the embryonic central nervous system suggest that the common, and therefore, ancestral function of Ptf1a/Fer1 is for the development of the CNS. In vertebrates, and likely in flies, this function involves a Notch-independent collaboration with Rbpj/Su(H) (H. Kei, Y. Nakada, S.M. Glasgow, T. Masui, M. Henke, T.M. Beres, M.A. Magnuson, R.J. MacDonald, and J.E. Johnson, in prep.). It would be of interest to determine whether RBPJ has Notch-independent roles in other programs of organogenesis with partners other than PTF1a.
Materials and methods
Recombinant DNA and cell transfection
Recombinant plasmids were constructed using standard molecular biological techniques. The promoter of mouse Rbpjl (−1380 to +103) was amplified by PCR from mouse BL6 genomic DNA and subcloned into pGL3b in front of the luciferase reporter gene (RbpLp.luc). The minimal promoter construct (EIp.luc) has the Ela1 basal promoter linked to the 5′ end of the luciferase gene of PGL3basic (Promega). Mutagenesis of PTF1-binding sites was performed with QuickChange (Stratagene); the E-boxes were modified from CANNTG to TCNNTA and the central cores of the TC-boxes from TTCCCA to TTCAAT. A repeat of three 32-bp PTF1-binding sites of Rbpjl promoter or three 26-bp PTF1 sites of the Ela1 promoter was placed upstream of EIp.luc. The cDNA expression plasmids for HEB (TCF12), mouse PTF1a (pcDNA3/ptf1a), mouse RBPJL (pcDNA3/Rbpjl), and myc-tagged human RBPJ have been described (Beres et al. 2006). Transfections of 293 human embryonic kidney cells (American Type Culture Collection [ATCC] CRL-1573), AR4-2J rat acinar cells (ATCC CRL-1492), and 266-6 mouse acinar cells (ATCC CRL-2151) were performed as described previously (Liu et al. 2001).
Quantitative RT–PCR
Complementary DNA was synthesized using SuperScript II reverse transcriptase (Invitrogen,) and oligo(dT) with RNA isolated from embryonic and adult mouse pancreas by the guanidine thiocyanate technique (MacDonald et al. 1987). Samples of cDNA derived from the equivalent of 15 ng of total cellular RNA template were amplified in 25-μL reactions with the following gene-specific primers: Ptf1a, 5′-CTTGCAGGGCACT CTCTTTC-3′ and 5′-CGATGTGAGCTGTCTCAGGA-3′; Rbpjl, 5′-CAGAGCATGCCATCATCCTA-3′ and 5′-AGTCCCATG TAACCGCAGAC-3′; Rbpj, 5′-GGTCCCAGACATTTCTGCAT-3′ and 5′-GGAGTTGGCTCTGAGAATCG-3′; Ela1, 5′-AGCAGA ACCTGAGCCAGAAT-3′ and 5′-TTGTTAGCCAGGATGGT TCC-3′; and β-actin, 5′-AGCCATGTACGTAGCCATCC-3′ and 5′-CTCTCAGCTGTGGTGGTGAA-3′. The amount of cDNA was quantified with SYBR Green MasterMix (Applied Biosystems) using the ABI prism 7700 (Applied Biosystems) and normalized to the level of β-actin.
Hybridization in situ
Hybridization in situ was performed with digoxygenin-labeled cRNA probes on frozen sections of paraformaldehyde-fixed mouse pancreas as described by others (Birren et al. 1993). Thirty-micron cryosections of mouse embryos were incubated with 1–2 μg/mL digoxygenin-labeled cRNA probes overnight at 65°C. The sections were then incubated with alkaline-phosphatase-conjugated anti-digoxygenin antibody and visualized with a 5-Bromo-4-Chloro-3′-Indolylphosphate p-Toluidine Salt and Nitro-Blue Tetrazolium Chloride solution. The mouse cRNA probes for Ptf1a, Rbpjl, and Ngn3 were synthesized with T7 or SP6 polymerase from PCR-amplified fragments of cDNA inserts cloned in pBSII SK+.
Electrophoretic mobility shift assays (EMSA)
The subunits of PTF1 were synthesized by in vitro transcription and translation using the TnT Reticulocyte Lysate System (Promega). Nuclear extract was prepared from rat adult pancreas as described previously (Rose et al. 2001). The products of IVT were quantified by [35S]methionine incorporation; relative molar ratios of the PTF1 subunits were calculated from the number of methionine residues in each. The double-stranded oligonucleotide probes for the PTF1-binding sites from the Rbpjl and the Ela1 promoter were labeled with 32P by polynucleotide kinase and EMSAs were performed as described (Rose et al. 1994).
Mouse genetic engineering
Generation and analysis of transient transgenic (G0) mouse embryos
The 1.3-Kb Rbpjl promoter from RbpLp.luc (see above) was ligated to a lacZ reporter gene in the pMCS5 cloning vector. The linear transgene DNA was separated from the plasmid vector and injected into the male pronucleus of fertilized eggs as described (Brinster et al. 1985). The eggs were cultured overnight and transferred to pseudopregnant ICR females. Embryos were obtained at E17.5, genotyped by PCR detection of the lacZ region, and stained for β-galactosidase activity as described previously (Hale et al. 2005).
Generation of mice bearing the Ptf1aWA allele
The single-codon W298A mutation was inserted into the endogenous Ptf1a locus of a mouse embryonic stem (ES) cell line by recombination-mediated cassette exchange (Feng et al. 1999) using a two-step staggered selection strategy (Long et al. 2004). A cassette acceptor allele of Ptf1a was engineered in R1 ES cells by a conventional homologous recombination approach and has the Ptf1a gene region between SalI sites at −1703 and +2398 replaced with a fragment containing neomycin resistance and thymidine kinase selection genes and bounded by inverted loxP sites. Construction of the cassette acceptor allele for the Ptf1a locus and details for the cassette exchange in ES cells will be described elsewhere (J. Burlison, Q. Long, C.V.E. Wright, and M.A. Magnuson, in prep.). The exchange cassette contained the Ptf1a gene SalI–SalI fragment from strain 129 genomic DNA with codon 298 tryptophan (TGG) changed to alanine (GCG) by PCR mutagenesis. The fragment was flanked by inverted loxP sites for the ES cell exchange reaction, and a thymidine kinase selection gene was attached outside the loxP sites. The exchange DNA, purified from the cloning vector, and a CMV-driven CRE expression plasmid were electroporated into the acceptor ES cell line described above, and the cultured cells were treated with hygromycin to select recombinants and with ganciclovir to select cells that had lost the thymidine kinase gene of the acceptor allele due to homologous replacement. The TGG-to-GCG change was confirmed in selected ES cell clones by DNA sequencing. Ptf1aWA/+ ES cells were injected into C57BL/6 blastocysts that were transferred to pseudopregnant ICR female mice. Male chimeras derived from the injected blastocysts were bred to C57BL/6 females and nonchimeric offspring were genotyped by a PCR protocol that distinguishes the 2-bp difference of the W298A allele. The Frt-flanked hygromycin resistance cassette was removed by crossing to a FlpE-expressing transgenic mouse (Rodriguez et al. 2000) prior to analysis.
Antibodies and immunofluorescence microscopy
For EMSA supershift experiments, the rabbit RBPJL antiserum and the affinity-purified rabbit anti-PTF1a have been described (Rose et al. 2001; Beres et al. 2006); anti-RBPJ was purchased from the Institute of Immunology Co., Ltd. For chromatin immunoprecipitation (ChIP), anti-RBPJ (SC-8213) and anti-cMYC were purchased from Santa Cruz Biotechnology. The anti-RBPJL did not cross-detect RBPJ, and the anti-RBPJ did not cross-detect RBPJL in EMSA and Western blotting experiments (Beres et al. 2006). For immunofluorescence, anti-CPA1 (used at 1:1000) and anti-PC1/3 (1:200) were from Chemicon, anti-insulin (1:2000) and anti-glucagon (1:8000) were from Linco, and anti-E-cadherin (1:1000) was from Sigma Aldrich. Affinity-purified Guinea pig anti-PTF1a (1:2000) for immunofluorescence was the kind gift of Drs. Kei Hori and Jane Johnson (University of Texas Southwestern Medical Center, Dallas, TX), and affinity-purified rabbit anti-PDX1 (1:1000) was a gracious gift from Michael Ray and Christopher Wright (Vanderbilt University School of Medicine, Nashville, TN). Immunofluorescent localization was performed with 10-μm frozen tissue sections derived from paraformaldehyde-fixed embryonic mouse pancreas. Staining with hematoxylin and Periodic Acid-Schiff was performed as described (Offield et al. 1996).
ChIP
Chromatin from adult mouse pancreas and liver was prepared from formaldehyde cross-linked nuclei as described for rat liver chromatin (Chaya and Zaret 2003), and sheared further to an average of 500-bp by sonication. Chromatin was prepared similarly from pooled embryonic pancreases, and the chromatin equivalent to that of one embryonic pancreas in 1 mL of ChIP dilution buffer (Upstate Biotechnology) was immunoprecipitated with 3 μg of antibody and blocked protein G-Sepharose beads (Upstate Biotechnology). After the beads were washed and cross-linking was reversed, the immunoprecipitated DNA was analyzed by quantitative real-time PCR. Sequential ChIP (Geisberg and Struhl 2004) was performed as described previously (Beres et al. 2006). The primer pairs to detect specific gene promoter regions were as follows: Rbpjl, 5′-TGCTGGGTCTG GCTTCTACT-3′ and 5′-CCGATCCTCACACTGGATTT-3′; Cpa1, 5′-CATGGTCAAGGGTGAAAGC-3′ and 5′-CTGAG GTCTGAGGCCTTTTT-3′; and R28S, 5′-CTGGGACATAGT GGGTGCTT-3′ and 5′-GAGCCTAGAGATGGGCTGTG-3′.
The amounts of immunoprecipitated DNA for the promoter regions were quantified with SYBR Green Mastermix (Applied Biosystems) using the ABI prism 7700 (Applied Biosystems). The level of ChIP enrichment was calculated from the amount of target promoter DNA relative to the amount of the 28S region of the large ribosomal RNA gene in the immunoprecipitated chromatin versus the starting chromatin.
Acknowledgments
We thank Mike Hale for assistance with the gene targeting constructs and many helpful discussions, Xuan Li for performing the gene targeting, Susan Hipkens for performing the RMCE, Jill Lindner for coordinating animal shipments and genotyping, Michael Ray and Chris Wright for PDX1 antibody, Kei Hori and Jane Johnson for the PTF1a antibody and insightful discussions, Victor Barbera for the in situ hybridizations, Jumin Xue for maintaining mouse lines, and Galvin Swift for advice, perspective, and critical readings of the manuscript. This study was supported by Public Health Service grants DK61220 (to R.J.M.) and DK42502 (to M.A.M.) and the β-Cell Biology Consortium of the National Institute of Diabetes and Digestive and Kidney Diseases.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1575207
References
- Ahlgren U., Jonsson J., Edlund H., Jonsson J., Edlund H., Edlund H. The morphogenesis of the pancreatic mesenchyme is uncoupled from that of the pancreatic epithelium in IPF1/PDX1-deficient mice. Development. 1996;122:1409–1416. doi: 10.1242/dev.122.5.1409. [DOI] [PubMed] [Google Scholar]
- Apelqvist A., Li H., Sommer L., Beatus P., Anderson D.J., Honjo T., de Hrabe Angelis M., Lendahl U., Edlund H., Li H., Sommer L., Beatus P., Anderson D.J., Honjo T., de Hrabe Angelis M., Lendahl U., Edlund H., Sommer L., Beatus P., Anderson D.J., Honjo T., de Hrabe Angelis M., Lendahl U., Edlund H., Beatus P., Anderson D.J., Honjo T., de Hrabe Angelis M., Lendahl U., Edlund H., Anderson D.J., Honjo T., de Hrabe Angelis M., Lendahl U., Edlund H., Honjo T., de Hrabe Angelis M., Lendahl U., Edlund H., de Hrabe Angelis M., Lendahl U., Edlund H., Lendahl U., Edlund H., Edlund H. Notch signalling controls pancreatic cell differentiation. Nature. 1999;400:877–881. doi: 10.1038/23716. [DOI] [PubMed] [Google Scholar]
- Beres T.M., Masui T., Swift G.H., Shi L., Henke R.M., MacDonald R.J., Masui T., Swift G.H., Shi L., Henke R.M., MacDonald R.J., Swift G.H., Shi L., Henke R.M., MacDonald R.J., Shi L., Henke R.M., MacDonald R.J., Henke R.M., MacDonald R.J., MacDonald R.J. PTF1 is an organ-specific and Notch-independent basic helix–loop–helix complex containing the mammalian Suppressor of Hairless (RBP-J) or its paralogue, RBP-L. Mol. Cell. Biol. 2006;26:117–130. doi: 10.1128/MCB.26.1.117-130.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birren S.J., Lo L., Anderson D.J., Lo L., Anderson D.J., Anderson D.J. Sympathetic neuroblasts undergo a developmental switch in trophic dependence. Development. 1993;119:597–610. doi: 10.1242/dev.119.3.597. [DOI] [PubMed] [Google Scholar]
- Brinster R.L., Chen H.Y., Trumbauer M.E., Yagle M.K., Palmiter R.D., Chen H.Y., Trumbauer M.E., Yagle M.K., Palmiter R.D., Trumbauer M.E., Yagle M.K., Palmiter R.D., Yagle M.K., Palmiter R.D., Palmiter R.D. Factors affecting the efficiency of introducing foreign DNA into mice by microinjecting eggs. Proc. Natl. Acad. Sci. 1985;82:4438–4442. doi: 10.1073/pnas.82.13.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaya D., Zaret K.S., Zaret K.S. Sequential chromatin immunoprecipitation from animal tissues. Methods Enzymol. 2003;376:361–372. doi: 10.1016/S0076-6879(03)76024-8. [DOI] [PubMed] [Google Scholar]
- Cockell M., Stevenson B.J., Strubin M., Hagenbuchle O., Wellauer P.K., Stevenson B.J., Strubin M., Hagenbuchle O., Wellauer P.K., Strubin M., Hagenbuchle O., Wellauer P.K., Hagenbuchle O., Wellauer P.K., Wellauer P.K. Identification of a cell-specific DNA-binding activity that interacts with a transcriptional activator of genes expressed in the acinar pancreas. Mol. Cell. Biol. 1989;9:2464–2476. doi: 10.1128/mcb.9.6.2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis C.A., Riddell D.C., Higgins M.J., Holden J.J.A., White B.N., Riddell D.C., Higgins M.J., Holden J.J.A., White B.N., Higgins M.J., Holden J.J.A., White B.N., Holden J.J.A., White B.N., White B.N. A gene family in Drosophila melanogaster coding for trypsin-like enzymes. Nucleic Acids Res. 1985;13:6605–6619. doi: 10.1093/nar/13.18.6605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esni F., Ghosh B., Biankin A.V., Lin J.W., Albert M.A., Yu X., MacDonald R.J., Civin C.I., Real F.X., Pack M.A., Ghosh B., Biankin A.V., Lin J.W., Albert M.A., Yu X., MacDonald R.J., Civin C.I., Real F.X., Pack M.A., Biankin A.V., Lin J.W., Albert M.A., Yu X., MacDonald R.J., Civin C.I., Real F.X., Pack M.A., Lin J.W., Albert M.A., Yu X., MacDonald R.J., Civin C.I., Real F.X., Pack M.A., Albert M.A., Yu X., MacDonald R.J., Civin C.I., Real F.X., Pack M.A., Yu X., MacDonald R.J., Civin C.I., Real F.X., Pack M.A., MacDonald R.J., Civin C.I., Real F.X., Pack M.A., Civin C.I., Real F.X., Pack M.A., Real F.X., Pack M.A., Pack M.A., et al. Notch inhibits Ptf1a function and acinar cell differentiation in developing mouse and zebrafish pancreas. Development. 2004a;131:4213–4224. doi: 10.1242/dev.01280. [DOI] [PubMed] [Google Scholar]
- Esni F., Stoffers D.A., Takeuchi T., Leach S.D., Stoffers D.A., Takeuchi T., Leach S.D., Takeuchi T., Leach S.D., Leach S.D. Origin of exocrine pancreatic cells from nestin-positive precursors in developing mouse pancreas. Mech. Dev. 2004b;121:15–25. doi: 10.1016/j.mod.2003.08.010. [DOI] [PubMed] [Google Scholar]
- Feng Y.Q., Seibler J., Alami R., Eisen A., Westerman K.A., Leboulch P., Fiering S., Bouhassira E.E., Seibler J., Alami R., Eisen A., Westerman K.A., Leboulch P., Fiering S., Bouhassira E.E., Alami R., Eisen A., Westerman K.A., Leboulch P., Fiering S., Bouhassira E.E., Eisen A., Westerman K.A., Leboulch P., Fiering S., Bouhassira E.E., Westerman K.A., Leboulch P., Fiering S., Bouhassira E.E., Leboulch P., Fiering S., Bouhassira E.E., Fiering S., Bouhassira E.E., Bouhassira E.E. Site-specific chromosomal integration in mammalian cells: Highly efficient CRE recombinase-mediated cassette exchange. J. Mol. Biol. 1999;292:779–785. doi: 10.1006/jmbi.1999.3113. [DOI] [PubMed] [Google Scholar]
- Fujikura J., Hosoda K., Iwakura H., Tomita T., Noguchi M., Masuzaki H., Tanigaki K., Yabe D., Honjo T., Nakao K., Hosoda K., Iwakura H., Tomita T., Noguchi M., Masuzaki H., Tanigaki K., Yabe D., Honjo T., Nakao K., Iwakura H., Tomita T., Noguchi M., Masuzaki H., Tanigaki K., Yabe D., Honjo T., Nakao K., Tomita T., Noguchi M., Masuzaki H., Tanigaki K., Yabe D., Honjo T., Nakao K., Noguchi M., Masuzaki H., Tanigaki K., Yabe D., Honjo T., Nakao K., Masuzaki H., Tanigaki K., Yabe D., Honjo T., Nakao K., Tanigaki K., Yabe D., Honjo T., Nakao K., Yabe D., Honjo T., Nakao K., Honjo T., Nakao K., Nakao K. Notch/Rbp-j signaling prevents premature endocrine and ductal cell differentiation in the pancreas. Cell Metab. 2006;3:59–65. doi: 10.1016/j.cmet.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Fujitani Y., Fujitani S., Boyer D.F., Gannon M., Kawaguchi Y., Ray M., Shiota M., Stein R.W., Magnuson M.A., Wright C.V.E., Fujitani S., Boyer D.F., Gannon M., Kawaguchi Y., Ray M., Shiota M., Stein R.W., Magnuson M.A., Wright C.V.E., Boyer D.F., Gannon M., Kawaguchi Y., Ray M., Shiota M., Stein R.W., Magnuson M.A., Wright C.V.E., Gannon M., Kawaguchi Y., Ray M., Shiota M., Stein R.W., Magnuson M.A., Wright C.V.E., Kawaguchi Y., Ray M., Shiota M., Stein R.W., Magnuson M.A., Wright C.V.E., Ray M., Shiota M., Stein R.W., Magnuson M.A., Wright C.V.E., Shiota M., Stein R.W., Magnuson M.A., Wright C.V.E., Stein R.W., Magnuson M.A., Wright C.V.E., Magnuson M.A., Wright C.V.E., Wright C.V.E. Targeted deletion of a cis-regulatory region reveals differential gene dosage requirements for Pdx1 in foregut organ differentiation and pancreas formation. Genes & Dev. 2006a;20:253–266. doi: 10.1101/gad.1360106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujitani Y., Fujitani S., Luo H., Qiu F., Burlison J., Long Q., Kawaguchi Y., Edlund H., MacDonald R.J., Furukawa T., Fujitani S., Luo H., Qiu F., Burlison J., Long Q., Kawaguchi Y., Edlund H., MacDonald R.J., Furukawa T., Luo H., Qiu F., Burlison J., Long Q., Kawaguchi Y., Edlund H., MacDonald R.J., Furukawa T., Qiu F., Burlison J., Long Q., Kawaguchi Y., Edlund H., MacDonald R.J., Furukawa T., Burlison J., Long Q., Kawaguchi Y., Edlund H., MacDonald R.J., Furukawa T., Long Q., Kawaguchi Y., Edlund H., MacDonald R.J., Furukawa T., Kawaguchi Y., Edlund H., MacDonald R.J., Furukawa T., Edlund H., MacDonald R.J., Furukawa T., MacDonald R.J., Furukawa T., Furukawa T., et al. Ptf1a determines horizontal and amacrine cell fates during mouse retinal development. Development. 2006b;133:4439–4450. doi: 10.1242/dev.02598. [DOI] [PubMed] [Google Scholar]
- Geisberg J.V., Struhl K., Struhl K. Quantitative sequential chromatin immunoprecipitation, a method for analyzing co-occupancy of proteins at genomic regions in vivo. Nucleic Acids Res. 2004;32:e151. doi: 10.1093/nar/gnh148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasgow S.M., Henke R.M., MacDonald R.J., Wright C.V.E., Johnson J.E., Henke R.M., MacDonald R.J., Wright C.V.E., Johnson J.E., MacDonald R.J., Wright C.V.E., Johnson J.E., Wright C.V.E., Johnson J.E., Johnson J.E. Ptf1a determines GABAergic over glutamatergic neuronal cell fate in the spinal cord dorsal horn. Development. 2005;132:5461–5469. doi: 10.1242/dev.02167. [DOI] [PubMed] [Google Scholar]
- Gradwohl G., Dierich A., LeMeur M., Guillemot F., Dierich A., LeMeur M., Guillemot F., LeMeur M., Guillemot F., Guillemot F. Neurogenin3 is required for the development of the four endocrine cell lineages of the pancreas. Proc. Natl. Acad. Sci. 2000;97:1607–1611. doi: 10.1073/pnas.97.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu G., Dubauskaite J., Melton D.A., Dubauskaite J., Melton D.A., Melton D.A. Direct evidence for the pancreatic lineage: NGN3+ cells are islet progenitors and are distinct from duct progenitors. Development. 2002;129:2447–2457. doi: 10.1242/dev.129.10.2447. [DOI] [PubMed] [Google Scholar]
- Hald J., Hjorth P., German M.S., Madsen O.D., Serup P., Jensen J., Hjorth P., German M.S., Madsen O.D., Serup P., Jensen J., German M.S., Madsen O.D., Serup P., Jensen J., Madsen O.D., Serup P., Jensen J., Serup P., Jensen J., Jensen J. Activated Notch1 prevents differentiation of pancreatic acinar cells and attenuate endocrine development. Dev. Biol. 2003;260:426–437. doi: 10.1016/s0012-1606(03)00326-9. [DOI] [PubMed] [Google Scholar]
- Hale M.A., Kagami H., Shi L., Holland A.M., Elsasser H.P., Hammer R.E., MacDonald R.J., Kagami H., Shi L., Holland A.M., Elsasser H.P., Hammer R.E., MacDonald R.J., Shi L., Holland A.M., Elsasser H.P., Hammer R.E., MacDonald R.J., Holland A.M., Elsasser H.P., Hammer R.E., MacDonald R.J., Elsasser H.P., Hammer R.E., MacDonald R.J., Hammer R.E., MacDonald R.J., MacDonald R.J. The homeodomain protein PDX1 is required at mid-pancreatic development for the formation of the exocrine pancreas. Dev. Biol. 2005;286:225–237. doi: 10.1016/j.ydbio.2005.07.026. [DOI] [PubMed] [Google Scholar]
- Jensen J., Pedersen E.E., Galante P., Hald J., Heller R.S., Ishibashi M., Kageyama R., Guillemot F., Serup P., Madsen O.D., Pedersen E.E., Galante P., Hald J., Heller R.S., Ishibashi M., Kageyama R., Guillemot F., Serup P., Madsen O.D., Galante P., Hald J., Heller R.S., Ishibashi M., Kageyama R., Guillemot F., Serup P., Madsen O.D., Hald J., Heller R.S., Ishibashi M., Kageyama R., Guillemot F., Serup P., Madsen O.D., Heller R.S., Ishibashi M., Kageyama R., Guillemot F., Serup P., Madsen O.D., Ishibashi M., Kageyama R., Guillemot F., Serup P., Madsen O.D., Kageyama R., Guillemot F., Serup P., Madsen O.D., Guillemot F., Serup P., Madsen O.D., Serup P., Madsen O.D., Madsen O.D. Control of endodermal endocrine development by Hes-1. Nat. Genet. 2000;24:36–44. doi: 10.1038/71657. [DOI] [PubMed] [Google Scholar]
- Jonsson J., Carlsson L., Edlund T., Edlund H., Carlsson L., Edlund T., Edlund H., Edlund T., Edlund H., Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371:606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- Kawaguchi Y., Cooper B., Gannon M., Ray M., MacDonald R.J., Wright C.V.E., Cooper B., Gannon M., Ray M., MacDonald R.J., Wright C.V.E., Gannon M., Ray M., MacDonald R.J., Wright C.V.E., Ray M., MacDonald R.J., Wright C.V.E., MacDonald R.J., Wright C.V.E., Wright C.V.E. The role of the transcriptional regulator PTF1a in converting intestinal to pancreatic progenitors. Nat. Genet. 2002;32:128–134. doi: 10.1038/ng959. [DOI] [PubMed] [Google Scholar]
- Kim S.K., MacDonald R.J., MacDonald R.J. Signaling and transcriptional control of pancreatic organogenesis. Curr. Opin. Genet. Dev. 2002;12:540–547. doi: 10.1016/s0959-437x(02)00338-6. [DOI] [PubMed] [Google Scholar]
- Kim S.K., Rulifson E.J., Rulifson E.J. Conserved mechanisms of glucose sensing and regulation by Drosophila corpora cardia cells. Nature. 2004;431:316–320. doi: 10.1038/nature02897. [DOI] [PubMed] [Google Scholar]
- Kovall R.A., Hendrickson W.A., Hendrickson W.A. Crystal structure of the nuclear effector of Notch signaling, CSL, bound to DNA. EMBO J. 2004;23:3441–3451. doi: 10.1038/sj.emboj.7600349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapp A., Knofler M., Frutiger F., Hughes G.J., Hagenbuchle O., Wellauer P.K., Knofler M., Frutiger F., Hughes G.J., Hagenbuchle O., Wellauer P.K., Frutiger F., Hughes G.J., Hagenbuchle O., Wellauer P.K., Hughes G.J., Hagenbuchle O., Wellauer P.K., Hagenbuchle O., Wellauer P.K., Wellauer P.K. The p48 DNA-binding subunit of transcription factor PTF1 is a new exocrine pancreas-specific basic helix–loop–helix protein. EMBO J. 1996;15:4317–4329. [PMC free article] [PubMed] [Google Scholar]
- Krapp A., Knofler M., Ledermann B., Burki K., Berney C., Zoerkler N., Hagenbuchle O., Wellauer P.K., Knofler M., Ledermann B., Burki K., Berney C., Zoerkler N., Hagenbuchle O., Wellauer P.K., Ledermann B., Burki K., Berney C., Zoerkler N., Hagenbuchle O., Wellauer P.K., Burki K., Berney C., Zoerkler N., Hagenbuchle O., Wellauer P.K., Berney C., Zoerkler N., Hagenbuchle O., Wellauer P.K., Zoerkler N., Hagenbuchle O., Wellauer P.K., Hagenbuchle O., Wellauer P.K., Wellauer P.K. The bHLH protein PTF1-p48 is essential for the formation of the exocrine and the correct spatial organization of the endocrine pancreas. Genes & Dev. 1998;12:3752–3763. doi: 10.1101/gad.12.23.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.C., Smith S.B., Watada H., Lin J., Scheel D., Wang J., Mirmira R.G., German M.S., Smith S.B., Watada H., Lin J., Scheel D., Wang J., Mirmira R.G., German M.S., Watada H., Lin J., Scheel D., Wang J., Mirmira R.G., German M.S., Lin J., Scheel D., Wang J., Mirmira R.G., German M.S., Scheel D., Wang J., Mirmira R.G., German M.S., Wang J., Mirmira R.G., German M.S., Mirmira R.G., German M.S., German M.S. Regulation of the pancreatic pro-endocrine gene neurogenin3. Diabetes. 2001;50:928–936. doi: 10.2337/diabetes.50.5.928. [DOI] [PubMed] [Google Scholar]
- Li H., Edlund H., Edlund H. Persistent expression of Hlxb9 in the pancreatic epithelium impairs pancreatic development. Dev. Biol. 2001;240:247–253. doi: 10.1006/dbio.2001.0440. [DOI] [PubMed] [Google Scholar]
- Liu Y., Swift G.H., MacDonald R.J., Swift G.H., MacDonald R.J., MacDonald R.J. The organization and function of the trimeric PDX1 homeodomain complex. J. Biol. Chem. 2001;276:17985–17993. doi: 10.1074/jbc.M100678200. [DOI] [PubMed] [Google Scholar]
- Long Q., Shelton K.D., Lindner J., Jones J.R., Magnuson M.A., Shelton K.D., Lindner J., Jones J.R., Magnuson M.A., Lindner J., Jones J.R., Magnuson M.A., Jones J.R., Magnuson M.A., Magnuson M.A. Efficient DNA cassette exchange in mouse embryonic stem cells by staggered positive–negative selection. Genesis. 2004;39:256–262. doi: 10.1002/gene.20053. [DOI] [PubMed] [Google Scholar]
- MacDonald R.J., Swift G.H., Przybyla A.E., Chirgwin J.M., Swift G.H., Przybyla A.E., Chirgwin J.M., Przybyla A.E., Chirgwin J.M., Chirgwin J.M. Isolation of RNA using guanidinium salts. Methods Enzymol. 1987;152:219–227. doi: 10.1016/0076-6879(87)52023-7. [DOI] [PubMed] [Google Scholar]
- Minoguchi S., Taniguchi Y., Kato H., Okazaki T., Strobl L.J., Zimber-Strobl U., Bornkamm G.W., Honjo T., Taniguchi Y., Kato H., Okazaki T., Strobl L.J., Zimber-Strobl U., Bornkamm G.W., Honjo T., Kato H., Okazaki T., Strobl L.J., Zimber-Strobl U., Bornkamm G.W., Honjo T., Okazaki T., Strobl L.J., Zimber-Strobl U., Bornkamm G.W., Honjo T., Strobl L.J., Zimber-Strobl U., Bornkamm G.W., Honjo T., Zimber-Strobl U., Bornkamm G.W., Honjo T., Bornkamm G.W., Honjo T., Honjo T. RBP-L, a transcription factor related to RBP-Jk. Mol. Cell. Biol. 1997;17:2679–2687. doi: 10.1128/mcb.17.5.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtaugh L.C., Stanger B.Z., Kwan D.M., Melton D.A., Stanger B.Z., Kwan D.M., Melton D.A., Kwan D.M., Melton D.A., Melton D.A. Notch signaling controls multiple steps of pancreatic differentiation. Proc. Natl. Acad. Sci. 2003;100:14920–14925. doi: 10.1073/pnas.2436557100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obata J., Yano M., Mimura H., Goto T., Nakayama R., Mibu Y., Oka C., Kawaichi M., Yano M., Mimura H., Goto T., Nakayama R., Mibu Y., Oka C., Kawaichi M., Mimura H., Goto T., Nakayama R., Mibu Y., Oka C., Kawaichi M., Goto T., Nakayama R., Mibu Y., Oka C., Kawaichi M., Nakayama R., Mibu Y., Oka C., Kawaichi M., Mibu Y., Oka C., Kawaichi M., Oka C., Kawaichi M., Kawaichi M. p48 subunit of mouse PTF1 binds to RBP-Jk/CBF1, the intracellular mediator of Notch signalling, and is expressed in the neural tube of early stage embryos. Genes Cells. 2001;6:345–360. doi: 10.1046/j.1365-2443.2001.00422.x. [DOI] [PubMed] [Google Scholar]
- Offield M.F., Jetton J.L., Labosky P.A., Ray M., Stein R.W., Magnuson M.A., Hogan B.L.M., Wright C.V.E., Jetton J.L., Labosky P.A., Ray M., Stein R.W., Magnuson M.A., Hogan B.L.M., Wright C.V.E., Labosky P.A., Ray M., Stein R.W., Magnuson M.A., Hogan B.L.M., Wright C.V.E., Ray M., Stein R.W., Magnuson M.A., Hogan B.L.M., Wright C.V.E., Stein R.W., Magnuson M.A., Hogan B.L.M., Wright C.V.E., Magnuson M.A., Hogan B.L.M., Wright C.V.E., Hogan B.L.M., Wright C.V.E., Wright C.V.E. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. 1996;122:983–995. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- Pictet R., Rutter W.J., Rutter W.J. Development of the embryonic endocrine pancreas. In: Steiner D.F., Freinkel N., Freinkel N., editors. Handbook of physiology section 7: Endocrinology. Williams and Wilkins; Baltimore, MD: 1972. pp. 25–66. [Google Scholar]
- Pictet R.L., Clark W.R., Williams R.H., Rutter W.J., Clark W.R., Williams R.H., Rutter W.J., Williams R.H., Rutter W.J., Rutter W.J. An ultrastructural analysis of the developing embryonic pancreas. Dev. Biol. 1972;29:436–467. doi: 10.1016/0012-1606(72)90083-8. [DOI] [PubMed] [Google Scholar]
- Rodriguez C.I., Bucholz F., Galloway J., Sequerra R., Kasper J., Ayala R., Stewart A.F., Dymecki S.M., Bucholz F., Galloway J., Sequerra R., Kasper J., Ayala R., Stewart A.F., Dymecki S.M., Galloway J., Sequerra R., Kasper J., Ayala R., Stewart A.F., Dymecki S.M., Sequerra R., Kasper J., Ayala R., Stewart A.F., Dymecki S.M., Kasper J., Ayala R., Stewart A.F., Dymecki S.M., Ayala R., Stewart A.F., Dymecki S.M., Stewart A.F., Dymecki S.M., Dymecki S.M. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat. Genet. 2000;25:139–140. doi: 10.1038/75973. [DOI] [PubMed] [Google Scholar]
- Rose S.D., MacDonald R.J., MacDonald R.J. Evolutionary silencing of the human elastase I gene (ELA1) Hum. Mol. Genet. 1997;6:897–903. doi: 10.1093/hmg/6.6.897. [DOI] [PubMed] [Google Scholar]
- Rose S.D., Kruse F., Swift G.H., MacDonald R.J., Hammer R.E., Kruse F., Swift G.H., MacDonald R.J., Hammer R.E., Swift G.H., MacDonald R.J., Hammer R.E., MacDonald R.J., Hammer R.E., Hammer R.E. A single element of the elastase I enhancer is sufficient to direct transcription selectively to the pancreas and gut. Mol. Cell. Biol. 1994;14:2048–2057. doi: 10.1128/mcb.14.3.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose S.D., Swift G.H., Peyton M.J., Hammer R.E., MacDonald R.J., Swift G.H., Peyton M.J., Hammer R.E., MacDonald R.J., Peyton M.J., Hammer R.E., MacDonald R.J., Hammer R.E., MacDonald R.J., MacDonald R.J. The role of PTF1-P48 in pancreatic acinar gene expression. J. Biol. Chem. 2001;276:44018–44026. doi: 10.1074/jbc.M106264200. [DOI] [PubMed] [Google Scholar]
- Roux E., Strubin M., Hagenbuchle O., Wellauer P.K., Strubin M., Hagenbuchle O., Wellauer P.K., Hagenbuchle O., Wellauer P.K., Wellauer P.K. The cell-specific transcription factor PTF1 contains two different subunits that interact with the DNA. Genes & Dev. 1989;3:1613–1624. doi: 10.1101/gad.3.10.1613. [DOI] [PubMed] [Google Scholar]
- Rulifson E.J., Kim S.K., Nusse R., Kim S.K., Nusse R., Nusse R. Ablation of insulin-producing neurons in flies: Growth and diabetic phenotypes. Science. 2002;296:1118–1120. doi: 10.1126/science.1070058. [DOI] [PubMed] [Google Scholar]
- Schwitzgebel V.M., Scheel D.W., Conners J.R., Kalamaras J., Lee J.E., Anderson D.J., Sussel L., Johnson J.D., German M.S., Scheel D.W., Conners J.R., Kalamaras J., Lee J.E., Anderson D.J., Sussel L., Johnson J.D., German M.S., Conners J.R., Kalamaras J., Lee J.E., Anderson D.J., Sussel L., Johnson J.D., German M.S., Kalamaras J., Lee J.E., Anderson D.J., Sussel L., Johnson J.D., German M.S., Lee J.E., Anderson D.J., Sussel L., Johnson J.D., German M.S., Anderson D.J., Sussel L., Johnson J.D., German M.S., Sussel L., Johnson J.D., German M.S., Johnson J.D., German M.S., German M.S. Expression of neurogenin3 reveals an islet cell precursor population in the pancreas. Development. 2000;127:3533–3542. doi: 10.1242/dev.127.16.3533. [DOI] [PubMed] [Google Scholar]
- Sellick G.S., Barker K.T., Stolte-Dijkstra I., Fleischmann C., Coleman R.J., Garrett C., Gloyn A.L., Edghill E.L., Hattersley A.T., Wellauer P.K., Barker K.T., Stolte-Dijkstra I., Fleischmann C., Coleman R.J., Garrett C., Gloyn A.L., Edghill E.L., Hattersley A.T., Wellauer P.K., Stolte-Dijkstra I., Fleischmann C., Coleman R.J., Garrett C., Gloyn A.L., Edghill E.L., Hattersley A.T., Wellauer P.K., Fleischmann C., Coleman R.J., Garrett C., Gloyn A.L., Edghill E.L., Hattersley A.T., Wellauer P.K., Coleman R.J., Garrett C., Gloyn A.L., Edghill E.L., Hattersley A.T., Wellauer P.K., Garrett C., Gloyn A.L., Edghill E.L., Hattersley A.T., Wellauer P.K., Gloyn A.L., Edghill E.L., Hattersley A.T., Wellauer P.K., Edghill E.L., Hattersley A.T., Wellauer P.K., Hattersley A.T., Wellauer P.K., Wellauer P.K., et al. Mutations in PTF1A cause pancreatic and cerebellar agenesis. Nat. Genet. 2004;36:1301–1305. doi: 10.1038/ng1475. [DOI] [PubMed] [Google Scholar]
- Wilson M.E., Kalamaras J.A., German M.S., Kalamaras J.A., German M.S., German M.S. Expression pattern of IAPP and prohormone convertase 1/3 reveals a distinctive set of endocrine cells in the embryonic pancreas. Mech. Dev. 2002;115:171–176. doi: 10.1016/s0925-4773(02)00118-1. [DOI] [PubMed] [Google Scholar]