Abstract
Why are most genes dispensable? The impact of gene deletions may depend on the environment (plasticity), the presence of compensatory mechanisms (mutational robustness), or both. Here, we analyze the interaction between these two forces by exploring the condition-dependence of synthetic genetic interactions that define redundant functions and alternative pathways. We performed systems-level flux balance analysis of the yeast (Saccharomyces cerevisiae) metabolic network to identify genetic interactions and then tested the model's predictions with in vivo gene-deletion studies. We found that the majority of synthetic genetic interactions are restricted to certain environmental conditions, partly because of the lack of compensation under some (but not all) nutrient conditions. Moreover, the phylogenetic cooccurrence of synthetically interacting pairs is not significantly different from random expectation. These findings suggest that these gene pairs have at least partially independent functions, and, hence, compensation is only a byproduct of their evolutionary history. Experimental analyses that used multiple gene deletion strains not only confirmed predictions of the model but also showed that investigation of false predictions may both improve functional annotation within the model and also lead to the discovery of higher-order genetic interactions. Our work supports the view that functional redundancy may be more apparent than real, and it offers a unified framework for the evolution of environmental adaptation and mutational robustness.
Keywords: epistasis, genetic robustness, Saccharomyces cerevisiae, environmental dependence, flux balance analysis
One of the most striking discoveries of molecular genetics is that a large fraction of the protein-coding genes have negligible effects on growth rates under standard laboratory conditions. Recent systematic single-gene-deletion studies suggest that nearly 80% of yeast genes appear not to be essential for growth (1). Comparable large-scale experiments in free-living bacteria, worm, and mouse showed that the fraction of essential genes is generally low, typically in the range of 6–19% (2, 3).
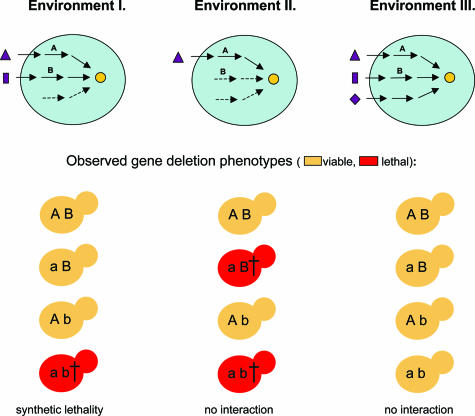
Although much investigated, the causes and evolution of gene dispensability remain controversial (4–7). The high fraction of dispensable genes might reflect the capacity of organisms to compensate for null mutations by using either redundant gene duplicates or alternative metabolic pathways (mutational robustness) (4). Others have suggested that many of the seemingly dispensable genes have important fitness contributions only under special environmental conditions (environmental adaptation) (5). However, the potential links between adaptation to new environmental conditions and robustness against harmful mutations have remained largely unexplored. It may well be that these theories on gene dispensability are not mutually exclusive. Differences in the availability of external nutrients and/or intracellular metabolites across environmental conditions can have a large effect on the number of active metabolic pathways that can produce a given key cellular component (Fig. 1). Hence, the capacity to compensate null mutations may vary substantially between different nutritional environments. One clear prediction of this idea is that the impact of both single- and double-gene deletions should change across environmental conditions.
Fig. 1.
Model to explain conditional synthetic lethality. A key metabolite (yellow circle) can be synthesized via three independent pathways. Metabolic genes A and B show synthetic lethality in Environment I, where starting nutrients of both pathways are present in the medium. However, B is unable to compensate deletion of A in Environment II, and the double mutant is rescued by the third pathway in Environment III.
Several lines of evidence are compatible with this idea. First, data compiled from available large-scale phenotypic screens in yeast [see supporting information (SI) Table 2] suggest that at least 20% of the ≈5,000 apparently nonessential genes in Saccharomyces cerevisiae make a large contribution to fitness under at least 1 of the 31 investigated conditions. Moreover, most of these conditionally essential genes make a contribution in only one or a few environments (Fig. 2), suggesting that conditional growth defects for numerous other gene deletions remain to be discovered. Second, a gene-deletion phenotype frequently does not reflect simply the absence of a given gene but also the response of the cell to its absence. Such responses may involve the redistribution of enzymatic fluxes in the network and up-regulation of previously inactive genes (8, 9). Third, mutagenesis studies on Escherichia coli and viruses have shown a joint influence of environmental plasticity and epistatic genetic interactions on the effect of deleterious mutations (10, 11).
Fig. 2.
Distribution of environmental specificity of single-gene deletion phenotypes. Gene deletions showing conditional growth phenotypes were compiled from published large-scale screens (see SI Table 2). Of 4,823 genes not essential for growth on YPD, 963 exhibited lethality or a strong growth defect under at least 1 of the 31 conditions investigated.
Using a combination of computational flux-balance analysis (FBA) and in vivo gene-deletion experiments, we have explored the link between epistatic genetic interactions and plasticity. FBA provides a rigorous computational framework for studying the impact of gene deletions (12). Based on steady-state assumptions and optimality criteria, this constraint-based method has been successfully applied for calculating the phenotypic behavior of the metabolic network (13) and the viability of single-gene-deletion strains in yeast (14). We restricted our attention to the most extreme form of genetic interaction [synthetic lethality (SL)], where a double deletant shows a no-growth phenotype that is not exhibited by either single deletant. The computational analyses suggest a strong dependence of genetic interactions on the prevailing environmental conditions, and this finding is supported by the experimental data presented below and by evidence from the literature.
Our study supports the view (15, 16) that mutational robustness is not a directly selected trait, but rather a byproduct of the evolution of biological networks toward survival under a wide range of environmental conditions (environmental robustness).
Results
FBA Reveals a High Frequency of Condition-Dependent Genetic Interactions.
We have extended previous studies (17) by applying FBA to a genome-scale metabolic network model of yeast (S. cerevisiae) to calculate genetic interactions. The previously reconstructed metabolic network (18) consists of 672 genes and 745 unique biochemical reactions and incorporates external nutrients and the corresponding transport processes. The impacts of all possible single- and double-gene deletions were calculated for 53 nutritional environments, including various carbon sources (see SI Materials and Methods). The analysis identified 98 gene pairs that were predicted to be involved in a SL relationship under at least one of the conditions investigated (SI Tables 3 and 4). Only 14.3% of these SL relationships were displayed under all nutrient conditions investigated, and 50% of them are restricted to only one or two nutritional environments (Fig. 3).
Fig. 3.
Distribution of environmental specificity of predicted synthetic lethal interactions. The histogram shows the distribution of the number of simulated environments where each of the 98 gene pairs exhibits synthetic lethality (only gene pairs interacting in at least 1 of the 53 conditions investigated are included).
The condition-specificity of interactions does not appear to be randomly distributed in the metabolic network. SL interactions between gene pairs annotated to different metabolic subsystems are present in a significantly smaller number of environments than those that are annotated to the same subsystem (Mann–Whitney U test, P < 0.02; because enzymes catalyzing the same reaction, by definition, have the same functional annotation, they were excluded from this analysis). Moreover, more than half (56.3 ± 2.7%) of all genetic interactions remain undetected when only a single environment is investigated. These results not only provide a link between compensation of null mutations and the environment but also suggest that systematic genetic interaction screens (which are generally restricted to a single condition) may miss many of the extant interactions.
Experimental Tests on the Reliability of the Model.
The predictions of the in silico model were tested by in vivo double-gene-deletion experiments (17 cases, Table 1, Materials and Methods) and by extracting published experimental data from the literature (32 cases; see SI Materials and Methods). This procedure enabled us to validate ≈60% of the total number of genetic interactions that we predicted to be present on either minimal or rich media (SI Table 5). Double deletants were constructed by sporulating and dissecting heterozygous diploids from crosses between two single-gene haploid deletants (see SI Materials and Methods). Next, we assessed the viability of double deletants by inspecting growth on plates. In 12 of the 17 cases investigated, we observed a clear synthetic sick or lethal (SSL) phenotype under the predicted growth condition (Table 1). The model also accurately captures changes in the presence of synthetic genetic interactions between media (see below). However, in five cases, the double mutant formed colonies qualitatively indistinguishable from the single-gene deletants.
Table 1.
Validation of in silico predictions by constructing double-mutant strains
| Gene 1 | Gene 2 | Environment | Prediction success | Measured epistasis |
|---|---|---|---|---|
| ASN1 | ASN2 | SD | + | SL |
| CHO2 | PCT1 | YPD | + | SS, −0.372* |
| CKI1 | CHO2 | YPD | − | −0.089* |
| CPT1 | CHO2 | YPD | − | 0.020 |
| ECM31 | FEN2 | YPD | + | SL |
| ECM31 | FEN2 | SD | + | SL |
| OPI3 | PCT1 | YPD | − | 0.012 |
| OPI3 | CPT1 | YPD | − | 0.004 |
| OPI3 | CKI1 | YPD | − | −0.067* |
| RPE1 | ZWF1 | YPD | + | SL |
| RPE1 | ZWF1 | SD | + | SL |
| SAM2 | SAM1 | YPD | + | SL |
| SAM2 | SAM1 | SD | + | SL |
| SPE1 | FEN2 | SD | + | SL |
| SPE2 | FEN2 | SD | + | SL |
| URA8 | URA7 | YPD | + | SL |
| URA8 | URA7 | SD | + | SL |
A set of SL interactions predicted for nutrient-rich (YPD) and/or glucose minimal (SD) media were validated by measuring the epistasis between pairs of gene deletions (see Materials and Methods). Lack of growth of a double mutant is denoted by “SL” and synthetic sickness by “SSA.” A prediction was considered successful if the double mutant had a visually apparent growth defect compared with single mutants in a plate growth assay (i.e. strong negative epistasis, SSL). ∗, P < 10−5.
These apparently false predictions may indicate that the model has only limited resolution. It may be, for example, that FBA accurately predicts the direction, but not the strength, of genetic interaction between genes. To explore whether weak genetic interactions, which are undetectable by a simple plate-growth assay, could be responsible for some of these false predictions, we measured the growth rates of all viable double deletants, and those of the corresponding single deletants, using an established protocol (19) (see Materials and Methods). In two of the five investigated cases, we found evidence for weak (but statistically significant) negative epistasis between the predicted gene pairs (Table 1).
Although only a limited number of interactions were tested experimentally, the results suggest that FBA can reliably detect genetic interactions in the metabolic network of yeast. Future large-scale experimental screens are required to get a precise estimate of the fraction of false-positive and false-negative predictions. As a preliminary to such a larger study, we augmented our experimental results with literature data available on single- and double-deletant strains (see SI Materials and Methods). Overall, we were able to test 49 predicted interactions (SI Table 5) and estimate that ≈49% (24 of 49) of them were correct and that, in 53% of the cases, at least the sign of epistasis was consistent with the predictions. In a similar vein, FBA can identify ≈24% of a curated list (20) of previously described SL interactions between metabolic genes. Both values are at least two orders of magnitude higher than expected by chance (P < 10−287, see SI Materials and Methods).
Gene Duplicates Can Explain Many of the False Predictions.
Lack of an observable growth defect in three of the experimentally observed cases could be due to the presence of gene duplicates with redundant functions that are not represented in the current metabolic reconstruction. We investigated this possibility by determining whether one or the other member of the gene pairs investigated had a gene duplicate that might provide compensation for one missing function in the double deletant. One member of the gene pair had a paralog in all three cases. Construction of triple-deletion strains (SI Materials and Methods) revealed strong negative epistasis in all three cases (SI Fig. 5).
For example, CHO2 and CPT1 are erroneously predicted to show a synthetic genetic interaction on rich medium. We hypothesized that this interaction might be masked by EPT1, a duplicate of CPT1. The two encoded proteins show 56% amino acid sequence similarity to each other and have different primary catalytic activities. However, some studies suggest that although Cpt1p accounts for 95% of phosphatidylcholine synthesis in vivo, the remaining 5% is likely to be catalyzed by Ept1p (21). Remarkably, deletion of all three genes simultaneously resulted in a much stronger growth defect than observed for any of the pair-wise deletions (SI Fig. 5).
In addition, our data suggest that interaction between OPI3 and PCT1 is masked by MUQ1, a distant paralog of PCT1 (the products of the two genes share 36% amino acid sequence similarity). The triple opi3Δ/pct1Δ/muq1Δ has a more severe phenotype than either double mutant (SI Fig. 5). Although Pct1p and Muq1p catalyze related reactions, they are generally believed to have different substrate specificities. Further biochemical studies will be needed to confirm whether Muq1p has the catalytic activity necessary to mitigate the effect of the pct1Δ/opi3Δ double deletion.
Detailed investigation of the false predictions can thus be used to generate novel biochemical hypotheses and refine the in silico model. Moreover, these results suggest that even duplicates with low sequence similarities and partly altered functions can compensate null mutations in each other. The importance of appropriate modeling of paralogs/gene duplicates is further underscored by inspection of false-negative predictions (i.e., true pair-wise interactions not predicted by the model). Many of these previously reported interacting gene pairs are predicted to participate in higher-order genetic interactions because of the presence of a gene duplicate with overlapping functions (i.e., an isoenzyme) (see SI Table 6). For example, TDH2 and TDH3 show synthetic lethality in vivo (22); however, our simulations show that tdh2Δ tdh3Δ double mutant is compensated by TDH1, a gene encoding an additional glyceraldehyde-3-phosphate dehydrogenase isoenzyme in the model. Lack of in vivo compensatory capacity of Tdh1p might be explained by its relatively low expression level compared with Tdh2p and Tdh3p (23). Thus, in addition to correctly assigning reactions to paralogous genes, incorporation of regulatory constraints (12) and information on maximum enzyme capacities would also be needed to more accurately model the behavior of isoenzymes. Transcriptional reprogramming upon gene deletion (8) may also have an influence on predicting deletant phenotypes.
Two Explanations for the Condition-Specificity of Genetic Interactions.
We found empirical evidence of condition-specific epistasis for 14 validated SSL gene pairs, of which 11 were correctly predicted (SI Table 7), suggesting that our modeling framework is able to capture variation in the incidence of genetic interactions across a range of environmental conditions. There could be at least two explanations for the condition-dependence of these genetic interactions (see Fig. 1). First, members of the synthetically interacting gene pairs make important individual contributions to growth under different nutritional conditions. Alternatively, the double-deletant strain becomes viable under different conditions. There is experimental evidence for both explanations (Fig. 4, SI Table 7).
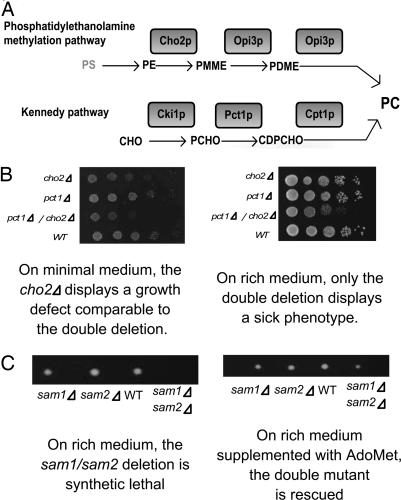
Fig. 4.
Examples of environment-specific synthetic genetic interactions. (A) Alternative routes to phosphatidylcholine biosynthesis in yeast. Cho2p, phosphatidylethanolamine methyltransferase; Opi3p, phospholipid methyltransferase; Cki1p, choline kinase; Pct1p, cholinephosphate cytidylyltransferase; Cpt1p, sn-1,2-diacylglycerol cholinephosphotransferase; PS, phosphatidylserine; PE, phosphatidylethanolamine; PME, phosphatidyl-N-methylethanolamine; PDME, phosphatidyl-N-dimethylethanolamine; CHO, choline; PCHO, choline phosphate; CDPCHO, CDP-choline; PC, phosphatidylcholine. (B) One member of the SSL pair makes an important individual contribution to growth under a different condition. CHO2 and PCT1 can compensate null mutations in one another under nutrient-rich (YPD) conditions, but the cho2Δ mutant is slow growing on minimal medium. (C) The double deletant becomes viable under a different condition. The SAM1/SAM2 duplicate gene pair, which encodes two distinct forms of S-adenosylmethionine (AdoMet) synthetase, can compensate null mutations in one another, and the double mutants are inviable under nutrient-rich (YPD) conditions. However, addition of AdoMet to the medium yields viable double mutants.
CHO2 and PCT1 are genes that encode two enzymes that each catalyze a step in two different pathways responsible for phosphatidylcholine synthesis (the phosphatidylethanolamine methylation pathway and the Kennedy pathway, respectively; see Fig. 4A). In agreement with our first explanation of condition-dependence, we find that these two genes can compensate null mutations in each other under nutrient-rich [yeast–peptone–dextrose (YPD)] conditions, but the cho2Δ deletant shows slow growth on glucose minimal medium (Fig. 4B). This indicates that, in the absence of exogenous choline, the Kennedy pathway (and hence PCT1), on its own, cannot support net phosphatidylcholine synthesis.
The second explanation for condition-dependence can be exemplified by the SAM1/SAM2 duplicate gene pair, which encodes two distinct forms of S-adenosylmethionine (AdoMet) synthetase. Although differentially regulated (24), the two genes can compensate null mutations in one another, and the double mutants are inviable under nutrient-rich (YPD) conditions (see Table 1). However, addition of AdoMet (the enzymatic product of Sam1p/Sam2p) into the medium yields viable double mutants (Fig. 4C).
Frequent Plasticity of Genetic Interactions Among Nonmetabolic Genes.
Having established the widespread occurrence of environment dependency of synthetic genetic interactions for metabolic genes, we asked whether condition dependency could be a general property of SSL interactions. First, we compiled a list of publicly available SSL interactions (25) discovered by a global genetic-interaction mapping approach (26) using a chemically defined glucose medium (those strains showing growth defects on minimal medium were excluded to ensure that the investigated interactions were not between unconditionally slow growing mutants, see Materials and Methods). Next, we collected viability data from published screens for single-gene-deletion phenotypes performed under 31 growth conditions (SI Table 2). In 57.4% of the investigated 2,666 SSL gene pairs, there is evidence that one or both members of the pairs make an essential contribution to growth under at least one of the 31 conditions investigated. This figure is likely to be an underestimate for two reasons: First, only a limited number of environments have been studied experimentally so far. Second, this estimate ignores cases where the double-deletant strain becomes viable under some other environmental condition. Moreover, there is some further support for a link between the extent of compensatory mechanisms and environmental specificity: genes for which evidence exists for conditional phenotypes have significantly more SL interactions than the rest of yeast's genes (Mann–Whitney U test, P = 0.002; see SI Fig. 6).
Random Phylogenetic Cooccurence of SSL Pairs.
Comparative genomics studies indicate that members of functional modules (i.e., genes that contribute jointly to a given cellular function) evolve nonindependently and show a similar phylogenetic distribution across species. For example, genes encoding members of protein complexes or metabolic modules are frequently gained and lost together during evolutionary history (27, 28). Indeed, we could confirm that gene pairs encoding subunits of the same literature-curated protein complexes (29) have higher phylogenetic cooccurrence than random gene pairs. We calculated a score (30) for the cooccurrence of these gene pairs across 16 eukaryotic genomes and used randomization protocols to get an estimate of statistical significance (see SI Materials and Methods). As expected, the score for subunits of protein complexes is significantly higher than expected by chance (P < 10−5, n = 7,186 pairs). Next, we asked if a similar result holds for experimentally determined SSL gene pairs. Using the same protocol as above, we found a strikingly different result. In contrast to members of protein complexes, gene pairs showing synthetic genetic interactions show no evidence for shared evolutionary history across species (P = 0.107, n = 1,850 pairs). Moreover, this finding cannot be explained by the likelihood of a low frequency of retention of redundant duplicates in all genomes; our result remains unchanged when all gene pairs showing even low sequence similarity to one another are excluded from the analysis (P = 0.113, n = 1,780 pairs, see SI Materials and Methods).
Discussion
Systematic screens on SL genetic interactions in yeast (25) and worm (31) are providing invaluable insights into the organisms' compensatory capacity. However, because of the enormous number of possible gene combinations, a complete mapping of SL interactions is still some way off. For this and other reasons, there is a need to find systems-biology models that are able to provide efficient and reliable tools for predicting (higher-order) genetic interactions. FBA offers a rigorous theoretical framework for studying the impact of multiple gene deletions on yeast metabolism. It also has a major advantage over other suggested computational approaches (32, 33) in that it can investigate epistasis under various environments.
Previous theoretical studies relied exclusively on the biochemical consistency of FBA to calculate epistasis (17, 34, 35). This study attempts to experimentally validate synthetic genetic interactions predicted by a genome-scale metabolic model. Although the accuracy of FBA at predicting genetic interactions is comparable with previous approaches (32, 33), the method is far from perfect. Our work suggests that many of the apparently false predictions are not due to major conceptual problems with FBA but, rather, are due to incomplete annotation and incorrect modeling of isozymes. First, a duplicate of a given enzyme-encoding gene could be present in the genome, which, although not annotated as an isozyme and diverged in both its amino acid sequence and biological function, could retain the ability to compensate for the absence of the other gene. Second, redundancy of certain isozymes annotated in the model might be more apparent than real because of incomplete compensatory capacity or regulatory differences between the gene copies (36). Thus, annotation of new enzymatic functions and incorporation of information on enzyme capacities and gene regulation (12) should lead to a refined model with more predictive power. Our study confirms the view that model building in systems biology is an iterative process (37) that proceeds by testing the predictions of the model against experimental data and then by using any discrepancies to revise and improve the model.
We used FBA to study the interplay between mutational robustness and the environment. Synthetic genetic interactions provide good examples of mutational robustness: members of these pairs are likely to be independent genes participating in alternative metabolic pathways or redundant gene duplicates. By integrating computational data with in vivo studies on double-gene deletants, we could show that synthetic genetic interactions are frequently restricted to particular environmental conditions, partly because genes involved in SL interactions under one condition frequently make an essential contribution to growth in another environment. The idea that compensating gene pairs bear distinct functional roles and are not redundant under all conditions is further supported by the observation that their phylogenetic cooccurrence is not different from those of functionally unrelated random gene pairs.
What could be the selective forces behind the evolutionary emergence of condition-specific compensation mechanisms? In principle, there are at least two possible routes. First, novel compensatory pathways might evolve to enhance robustness against spontaneously arising deleterious mutations and may later provide raw material for adaptation to new environments (38). Alternatively, adaptation toward new nutritional conditions may drive the evolution of novel metabolic pathways and, as a correlated response, some of these new pathways may also enhance the organism's ability to withstand harmful mutations under certain conditions. For example, we speculate that the ancestor of the choline transporter gene (HNM1) might have evolved to enable the cell to use exogenous choline and, as a side effect, provides robustness against null mutations in genes of the phosphatidylethanolamine methylation pathway when choline is present in the medium.
Several lines of theoretical reasoning and observation are consistent with the view that mutational robustness is a byproduct of other evolved properties of metabolic networks. First, the presence of compensating metabolic gene duplicates can be explained by gene dosage effects (5), differential regulation (39), or the capacity to filter nonheritable noise (40), without the need to invoke direct selection to favor mutational resilience. In a similar vein, computer simulations suggest that the evolution of several structural properties of metabolic networks can be explained by selection for enhanced growth rates (41). Second, population-genetics models have clearly shown that the selection pressure for enhanced mutational robustness is generally weak, of the order of mutation rates (42). Similar objections were raised to Fisher's selectionist theory of dominance (43). In contrast, evolution of environmental robustness is unproblematic from a population genetics point of view (42, 44), and mutational robustness might simply arise as a correlated response to selection for environmental robustness (15, 16). The finding that the extent of epistatic interactions is not independent of environmental specificity (SI Fig. 6 and ref. 10) provides evidence for a correlation between mutational and environmental robustness. Finally, the scenario of direct selection for mutational robustness would leave unexplained our observation that different genes can be compensated in different environments. Therefore, based on the above arguments, we conclude that mutational robustness of metabolic networks is unlikely to be a directly selected trait. Rather, it is a side effect of adaptation to survive in a large variety of nutrient conditions.
Materials and Methods
Analysis of Genetic Interactions in the Metabolic Network of Yeast.
We examined a recently updated (iLL672) metabolic network of S. cerevisiae, which contains 672 genes and 745 unique biochemical reactions including transport processes (18). The reconstruction also provides information on the association of genes with different metabolic subsystems (e.g., purine metabolism, phospholipid biosynthesis, etc.). One dubious reaction, corresponding to choline biosynthesis, was removed from the reconstruction because yeasts are unable to synthesize choline de novo (45). FBA of the metabolic network was used to calculate the impact of gene deletions on maximum biomass production rate (a proxy for fitness). Details of the FBA protocol have been described in ref. 12. SL interactions were identified by simulating all possible single- and double-gene deletions and screening for gene pairs where the single deletions had a <10% fitness effect, but the double mutant was unable to produce biomass (the use of different cut-offs led to very similar results). All deletion simulations were carried out in the ura3Δ leu2Δ his3Δ met17Δ lys2Δ genetic background to most closely mimic the strains used in the in vivo studies (see SI Materials and Methods).
To explore the condition dependency of SL interactions, we defined a large set of nutrient environments. First, we tested all external nutrients for their ability to support aerobic growth in minimal medium. This resulted in 50 minimal media containing different principal carbon sources, including glucose. Additionally, we defined a medium mimicking YPD, a medium where all possible external nutrients were allowed for uptake, and a minimal vitamin medium [lacking pantothenate because yeast is capable of de novo pantothenate biosynthesis (46)], resulting in 53 environmental conditions (for details see SI Materials and Methods and SI Table 8). All simulated growth media were supplemented with uracil, leucine, histidine, methionine, and lysine to complement the nutritional markers and also with vitamins (with or without pantothenate, see above) to further mimic the experimental conditions.
Experimental Procedures.
The simulations identified 59 gene pairs showing SL on either nutrient-rich (YPD) or glucose minimal [synthetic defined (SD)] medium. Published data (1, 18) on single-deletion phenotypes for these two conditions enabled us, in a comprehensive manner, to identify gene pairs for which the viability of single deletants was correctly predicted.
To carry out in vivo validation, we considered initially those gene pairs that have, at most, one paralog that is not annotated as an isozyme in the model. This choice enabled us to test higher-order genetic interactions by constructing triple-deletion strains in cases where the double mutant was viable (see below). Because, by using this criterion, all isozymes were excluded from validation, we additionally incorporated three randomly selected isozyme pairs in our experimental set. Moreover, among the group of gene pairs containing several paralogs, we decided to test gene pairs involved in pantothenate and polyamine biosynthesis for which we had the highest number of predicted SL interactions but no literature support available. Finally, some of the selected gene pairs could not be verified because one or the other mutant strain was missing from the deletion collection or contained a second-site mutation (18). This selection procedure left us with a set of 13 gene pairs to validate in vivo. These pairs corresponded to 17 cases of synthetic lethality: 6 on YPD, 3 on glucose minimal medium (SD), and 4 on both media.
We constructed the predicted double mutants by crossing haploid yeast strains containing single-gene deletions in the BY4742 and BY4741 backgrounds following standard protocols (see SI Materials and Methods).
In cases where we failed to detect any growth defect by visual inspection of plates (no overt synthetic sick or lethal phenotype), we performed accurate measurements of maximum growth rates of single and double mutants to estimate epistasis. Optical densities were measured by a Bioscreen C analyzer (Thermic Labsystems, Oy, Finland), and maximum growth rates were calculated by using an established protocol (19). Five cultures were grown for each strain in both YPD and SD. Maximum growth rates were averaged over the five replicates and divided by the wild-type value to yield a relative growth rate for each strain. Because additivity of the growth rates is equivalent to multiplicity of nonlogarithmic measures of fitness (47), we defined epistasis (ε) as the degree of departure from additivity of the relative growth rates (μ), thus ε = μAB + μab − μAb − μaB.
See SI Materials and Methods for details on the construction of triple-gene deletants.
Analysis of Global Genetic Interaction and Mutant Phenotype Data Sets.
We compiled a list of publicly available SSL interactions (25), discovered by the synthetic genetic array approach (26), on SD medium complemented with amino acids. Single-gene deletion strains that exhibit a pronounced growth defect (<80% of wild-type growth rate) on SD medium (48) were excluded from further analyses to ensure that the interactions were not between unconditionally slow growing mutants. This resulted in a list of 2,666 synthetic genetic interactions (1,230 of them being SL). Information on environment-specific phenotypes of single-gene deletions in nonessential genes was collected from published large-scale phenotypic screens (see SI Table 2). Our list of 31 experimental conditions included various nutrient and stress conditions, sporulation, and stationary phase, but excluded drug treatments. Only the strongest growth defects and phenotypes were considered as evidence for conditional fitness contributions.
Supplementary Material
Acknowledgments
We thank Lars Blank and Uwe Sauer for providing early access to the iLL672 metabolic reconstruction and to data from phenotypic screening of single mutants. We acknowledge valuable comments from Jonas Warringer on the growth-rate measurement protocol. R.H. was supported by a Biotechnology and Biological Sciences Research Council (BBSRC) studentship. C.P. and B.P. are supported by the Hungarian Scientific Research Fund. B.P. is a Long-Term Fellow of The International Human Frontier Science Program Organization, C.P. is a Long-Term Fellow of the European Molecular Biology Organization, and D.D. is a Natural Environment Research Council (U.K.) Advanced Fellow. Work on systems biology in S.G.O.'s laboratory was supported by BBSRC. We acknowledge support from a Royal Society research grant (to D.D.).
Abbreviations
- FBA
flux-balance analysis
- SD
synthetic defined
- SL
synthetic lethal
- SS
synthetic sickness
- SSL
synthetic sick/lethal
- YPD
yeast–peptone–dextrose.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0607153104/DC1.
References
- 1.Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, et al. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 2.Koonin EV. Nat Rev Microbiol. 2003;1:127–136. doi: 10.1038/nrmicro751. [DOI] [PubMed] [Google Scholar]
- 3.Wilson L, Ching YH, Farias M, Hartford SA, Howell G, Shao H, Bucan M, Schimenti JC. Genome Res. 2005;15:1095–1105. doi: 10.1101/gr.3826505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wagner A. Nat Genet. 2000;24:355–361. doi: 10.1038/74174. [DOI] [PubMed] [Google Scholar]
- 5.Papp B, Pál C, Hurst LD. Nature. 2004;429:661–664. doi: 10.1038/nature02636. [DOI] [PubMed] [Google Scholar]
- 6.Blank LM, Kuepfer L, Sauer U. Genome Biol. 2005;6:R49. doi: 10.1186/gb-2005-6-6-r49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gu Z, Steinmetz LM, Gu X, Scharfe C, Davis RW, Li WH. Nature. 2003;421:63–66. doi: 10.1038/nature01198. [DOI] [PubMed] [Google Scholar]
- 8.Kafri R, Bar-Even A, Pilpel Y. Nat Genet. 2005;37:295–299. doi: 10.1038/ng1523. [DOI] [PubMed] [Google Scholar]
- 9.Fischer E, Sauer U. Eur J Biochem. 2003;270:880–891. doi: 10.1046/j.1432-1033.2003.03448.x. [DOI] [PubMed] [Google Scholar]
- 10.Remold SK, Lenski RE. Nat Genet. 2004;36:423–426. doi: 10.1038/ng1324. [DOI] [PubMed] [Google Scholar]
- 11.You L, Yin J. Genetics. 2002;160:1273–1281. doi: 10.1093/genetics/160.4.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Price ND, Reed JL, Palsson BO. Nat Rev Microbiol. 2004;2:886–897. doi: 10.1038/nrmicro1023. [DOI] [PubMed] [Google Scholar]
- 13.Famili I, Forster J, Nielsen J, Palsson BO. Proc Natl Acad Sci USA. 2003;100:13134–13139. doi: 10.1073/pnas.2235812100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forster J, Famili I, Palsson BO, Nielsen J. Omics. 2003;7:193–202. doi: 10.1089/153623103322246584. [DOI] [PubMed] [Google Scholar]
- 15.Wagner A. Robustness and Evolvability of Living Systems. Princeton: Princeton Univ Press; 2005. [Google Scholar]
- 16.Gibson G, Wagner G. BioEssays. 2000;22:372–380. doi: 10.1002/(SICI)1521-1878(200004)22:4<372::AID-BIES7>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 17.Segrè D, Deluna A, Church GM, Kishony R. Nat Genet. 2005;37:77–83. doi: 10.1038/ng1489. [DOI] [PubMed] [Google Scholar]
- 18.Kuepfer L, Sauer U, Blank LM. Genome Res. 2005;15:1421–1430. doi: 10.1101/gr.3992505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Warringer J, Blomberg A. Yeast. 2003;20:53–67. doi: 10.1002/yea.931. [DOI] [PubMed] [Google Scholar]
- 20.Reguly T, Breitkreutz A, Boucher L, Breitkreutz BJ, Hon GC, Myers CL, Parsons A, Friesen H, Oughtred R, Tong A, et al. J Biol. 2006;5:11. doi: 10.1186/jbiol36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McMaster CR, Bell RM. J Biol Chem. 1994;269:28010–28016. [PubMed] [Google Scholar]
- 22.McAlister L, Holland MJ. J Biol Chem. 1985;260:15013–15018. [PubMed] [Google Scholar]
- 23.McAlister L, Holland MJ. J Biol Chem. 1985;260:15019–15027. [PubMed] [Google Scholar]
- 24.Thomas D, Rothstein R, Rosenberg N, Surdin-Kerjan Y. Mol Cell Biol. 1988;8:5132–5139. doi: 10.1128/mcb.8.12.5132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tong AH, Lesage G, Bader GD, Ding H, Xu H, Xin X, Young J, Berriz GF, Brost RL, Chang M, et al. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- 26.Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, Page N, Robinson M, Raghibizadeh S, Hogue CW, Bussey H, et al. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- 27.Pál C, Papp B, Lercher MJ. Nat Genet. 2005;37:1372–1375. doi: 10.1038/ng1686. [DOI] [PubMed] [Google Scholar]
- 28.Campillos M, von Mering C, Jensen LJ, Bork P. Genome Res. 2006;16:374–382. doi: 10.1101/gr.4336406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Güldener U, Münsterkötter M, Kastenmüller G, Strack N, van Helden J, Lemer C, Richelles J, Wodak SJ, García-Martínez J, Pérez-Ortín JE, et al. Nucleic Acids Res. 2005;33:D364–D368. doi: 10.1093/nar/gki053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huynen M, Snel B, Lathe W, III, Bork P. Genome Res. 2000;10:1204–1210. doi: 10.1101/gr.10.8.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lehner B, Crombie C, Tischler J, Fortunato A, Fraser AG. Nat Genet. 2006;38:896–903. doi: 10.1038/ng1844. [DOI] [PubMed] [Google Scholar]
- 32.Wong SL, Zhang LV, Tong AH, Li Z, Goldberg DS, King OD, Lesage G, Vidal M, Andrews B, Bussey H, et al. Proc Natl Acad Sci USA. 2004;101:15682–15687. doi: 10.1073/pnas.0406614101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhong W, Sternberg PW. Science. 2006;311:1481–1484. doi: 10.1126/science.1123287. [DOI] [PubMed] [Google Scholar]
- 34.Thiele I, Vo TD, Price ND, Palsson BO. J Bacteriol. 2005;187:5818–5830. doi: 10.1128/JB.187.16.5818-5830.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ghim CM, Goh KI, Kahng B. J Theor Biol. 2005;237:401–411. doi: 10.1016/j.jtbi.2005.04.025. [DOI] [PubMed] [Google Scholar]
- 36.Delneri D, Gardner DCJ, Oliver SG. Genetics. 1999;153:1591–1600. doi: 10.1093/genetics/153.4.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Covert MW, Knight EM, Reed JL, Herrgard MJ, Palsson BO. Nature. 2004;429:92–96. doi: 10.1038/nature02456. [DOI] [PubMed] [Google Scholar]
- 38.Deutscher D, Meilijson I, Kupiec M, Ruppin E. Nat Genet. 2006;38:993–998. doi: 10.1038/ng1856. [DOI] [PubMed] [Google Scholar]
- 39.Ihmels J, Levy R, Barkai N. Nat Biotechnol. 2004;22:86–92. doi: 10.1038/nbt918. [DOI] [PubMed] [Google Scholar]
- 40.Kafri R, Levy M, Pilpel Y. Proc Natl Acad Sci USA. 2006;103:11653–11658. doi: 10.1073/pnas.0604883103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pfeiffer T, Soyer OS, Bonhoeffer S. PLoS Biol. 2005;3:e228. doi: 10.1371/journal.pbio.0030228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Proulx SR. Am Nat. 2005;165:147–162. doi: 10.1086/426873. [DOI] [PubMed] [Google Scholar]
- 43.Wright S. Am Nat. 1929;63:274–279. [Google Scholar]
- 44.Wagner GP, Booth G, Bagheri-Chaichian H. Evol Int J Org Evol. 1997;51:329–347. doi: 10.1111/j.1558-5646.1997.tb02420.x. [DOI] [PubMed] [Google Scholar]
- 45.Howe AG, Zaremberg V, McMaster CR. J Biol Chem. 2002;277:44100–44107. doi: 10.1074/jbc.M206643200. [DOI] [PubMed] [Google Scholar]
- 46.White WH, Gunyuzlu PL, Toyn JH. J Biol Chem. 2001;276:10794–10800. doi: 10.1074/jbc.M009804200. [DOI] [PubMed] [Google Scholar]
- 47.Szafraniec K, Wloch DM, Sliwa P, Borts RH, Korona R. Genet Res. 2003;82:19–31. doi: 10.1017/s001667230300630x. [DOI] [PubMed] [Google Scholar]
- 48.Warringer J, Ericson E, Fernandez L, Nerman O, Blomberg A. Proc Natl Acad Sci USA. 2003;100:15724–15729. doi: 10.1073/pnas.2435976100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.