Abstract
The identification of regulatory regions is one of the most important and challenging problems toward the functional annotation of the human genome. In higher eukaryotes, transcription-factor (TF) binding sites are often organized in clusters called cis-regulatory modules (CRM). While the prediction of individual TF-binding sites is a notoriously difficult problem, CRM prediction has proven to be somewhat more reliable. Starting from a set of predicted binding sites for more than 200 TF families documented in Transfac, we describe an algorithm relying on the principle that CRMs generally contain several phylogenetically conserved binding sites for a few different TFs. The method allows the prediction of more than 118,000 CRMs within the human genome. A subset of these is shown to be bound in vivo by TFs using ChIP-chip. Their analysis reveals, among other things, that CRM density varies widely across the genome, with CRM-rich regions often being located near genes encoding transcription factors involved in development. Predicted CRMs show a surprising enrichment near the 3′ end of genes and in regions far from genes. We document the tendency for certain TFs to bind modules located in specific regions with respect to their target genes and identify TFs likely to be involved in tissue-specific regulation. The set of predicted CRMs, which is made available as a public database called PReMod (http://genomequebec.mcgill.ca/PReMod), will help analyze regulatory mechanisms in specific biological systems.
The regulation of gene expression is at the core of many important biological processes such as cell growth, division, differentiation, and adaptation to the extracellular environment. Gene expression is regulated in large part at the transcription level, with transcription factors (TFs) binding their specific DNA regulatory elements and activating or repressing transcription. The identification and characterization of these DNA regulatory elements are among the most important and challenging tasks for molecular biologists in the post-genome era.
TFs typically have an affinity for short, 5–15 bp, degenerate DNA sequences. Decades of work in many laboratories have led to the identification of consensus-binding motifs for hundreds of these TFs. These binding motifs are generally represented by position-weighted matrices (PWM). In principle, examination of the human genome with these PWM should allow for the identification of TF-binding sites (TFBSs), and hence, regulatory regions; but the size of the genome, combined with the fact that TF-binding motifs are short and degenerate, complicates this task enormously. Indeed, these motifs can be found everywhere in the genome and experiments have shown that only an extremely small proportion represent bona fide TFBSs. The binding of a TF is thus not simply a function of the theoretical affinity for a DNA site, but also of a number of other factors like the chromatin environment and the cooperation or competition with other DNA-binding proteins. In higher eukaryotes, TFs rarely operate by themselves, but rather bind to DNA in cooperation with other DNA-binding proteins. The DNA footprint of this set of factors is called a cis-regulatory module (CRM), which consists of a set of TFBSs located in a DNA region of up to a few hundred bases located in the vicinity of the gene being regulated. These modules have been the focus of much work recently (Davidson 2001), particularly in the context of the gene regulation during development (Howard and Davidson 2004), and are believed to be key features of most transcriptional regulatory processes in mammals.
Several features of known CRMs can be used to recognize new modules as follows: (1) CRMs are generally composed of several binding sites for a few different TFs; (2) CRMs, and in particular the binding sites they contain, are generally more evolutionarily conserved than their flanking intergenic regions, and (3) genes regulated by a common set of TFs tend to be coexpressed. Different combinations of those characteristics have been used, often in conjunction with PWM information, to predict regulatory elements for specific TFs. However, very few existing methods are designed to be applied on a genome-wide scale without prior knowledge about sets of interacting TFs or sets of coregulated genes (the main exception being the regulatory potential analysis of Kolbe et al. [2004] and King et al. [2005]). To date, the general properties of human nonpromoter regulatory regions indeed remain largely unexplored.
Here, we describe an algorithm that allows the identification of about 118,000 putative CRMs, based on predicted sites of 229 families of human TFs (represented by 481 PWMs). We refer to these regions as ‘‘predicted cis-regulatory modules’’ (pCRMs). Together with the regions predicted for regulatory potential by Kolbe et al. (2004), this constitutes the first genome-wide, nonpromoter centric set of human cis-regulatory modules, although related studies have been reported for yeast (Segal et al. 2003) and for human promoters (Bajic et al. 2004; Segal and Sharan 2005; Robertson et al. 2006). More importantly, in the analysis our set of pCRMs yields a number of novel insights into the mechanisms of gene regulation. After experimental validation of some of our predictions using a combination of chromatin immunoprecipitation and DNA microarrays (ChIP-chip), we used these predictions to explore the regulatory potential of the human genome. We show that, despite the fact that our pCRMs undoubtedly contain a significant number of false positives, the whole-genome approach provides sufficient statistical power to formulate specific biological hypotheses. For example, (1) the CRM density is unexpectedly high downstream of the 3′ end of genes, hinting at a possible involvement in regulating antisense transcription; (2) the regions that are the densest in CRMs are associated with developmental TFs; (3) different TF families have binding sites that are enriched in different regions relative to their target genes; (4) certain TFs or combination of TFs are associated with tissue-specific regulation. The Web-accessible database that accompanies this study will prove useful to experimental biologists interested in the regulation of specific genes, and will allow further bioinformatics and data-mining efforts.
Results and Discussion
Existing methods for cis-regulatory module prediction
The problem of computationally predicting cis-regulatory modules has been extensively studied in the last few years. Most predictive methods are either based exclusively on sequence data (see below), but some attempt to take advantage of gene expression data (Segal et al. 2003; Ihmels et al. 2004; Kloster et al. 2005; Wang et al. 2005) or DNaseI hypersensitivity data (Noble et al. 2005). Sequence-based algorithms have been developed along several lines. In the most studied case, the promoters of a set of (presumably) coregulated genes obtained from some prior experiments is analyzed to identify overrepresented motif combinations likely to be responsible for the gene’s coregulation (Wasserman and Fickett 1998; Krivan and Wasserman 2001; Aerts et al. 2003, 2004; Sharan et al. 2004; Thompson et al. 2004; Zhou and Wong 2004; Gupta and Liu 2005; Segal and Sharan 2005). Other approaches assume that the user provides a small set of transcription-factor PWMs that are expected to co-occur in modules, and identifies genomic regions densely populated in putative sites for these TFs (Bailey and Noble 2003; Frith et al. 2003; Johansson et al. 2003; Sinha et al. 2003, 2004; Alkema et al. 2004). None of these two families of approaches are applicable in our setting, where we do not have sets of coregulated genes to train from, and where we have little prior knowledge about combinations of factors that are likely to co-occur to form modules.
To our knowledge, the only computational approach that has been used for de novo, genome-wide prediction of regulatory regions is the method of regulatory potential estimation from Hardison’s group (Kolbe et al. 2004; King et al. 2005). This method is trained to recognize sequence features and interspecies conservation patterns that allow us to distinguish between known regulatory regions and nonfunctional sequences. A comparison of the results obtained by this approach and ours is given below.
A new algorithm for prediction of cis-regulatory modules
We designed a computational method with the goal of (1) identifying the DNA regions within the human genome that are likely to be important for regulating gene expression and (2) predicting what TFs are likely to bind these regions. Because our interest does not lie on any specific TF or specific system, but rather on having a global map of the regulatory elements of the entire genome, we exploited the fact that PWMs representing binding sites for a few hundreds of TFs have been described in databases such as Transfac (Matys et al. 2003) and JASPAR (Sandelin et al. 2004). Our algorithm takes advantage of the fact that regulatory regions often consist of clusters of binding sites for a few different TFs and that they are more conserved than their flanking intergenic DNA (Davidson 2001; Bulyk 2003; Levine and Tjian 2003). Our approach, based on the detection of statistically significant clusters of phylogenetically conserved TFBSs, shares some of the features of algorithms previously proposed by Sharan et al. (2004) and Aerts et al. (2004), but differs in that it allows the detection of modules without prior knowledge regarding which TFs are likely to be involved together in modules of interest. Our method also shares some similarities with the word-based approach of Philipakis et al. (2005), but uses a very different approach to module scoring.
Our algorithm involves two steps (see Fig. 1 and Methods for more details) as follows:
Figure 1.
Overview of the CRM prediction algorithm. TFBS predictions for different PWMs are shown with different geometric shapes and their size indicates the score of the hit. Hits from individual species are combined using a weighted average method to compute the “Aligned hits.” The most significant (up to five) aligned hits are considered as “Tags” for the corresponding region. The sum of the Tags scores is used to calculate a “Module score” using a statistical significance estimation. This operation is performed for each position of the human genome, for sliding windows of size 100, 200, 500, 1000, and 2000 bp.
Identification and scoring of putative TFBSs using 481 Transfac PWMs for vertebrate TFs (representing a total of 229 TF families). To this end, each noncoding, nonrepetitive position of the human genome within a human–mouse–rat alignment block (based on MULTIZ genome-wide alignments [Blanchette et al. 2004]) was evaluated for its similarity to each PWM using a log-likelihood ratio score with a third-order Markov background model parameterized based on the local GC content. Corresponding orthologous positions in mouse and rat genomes were evaluated similarly and a weighted average of the human, mouse, and rat log-likelihood scores at aligned positions was used to define a ‘‘hit score’’ for each human genomic position and each PWM. The scoring method favors simultaneous matches in all three species, which greatly reduces the false-positive rate of predictions. Notice, however, that the sites predicted need not be located within large phylogenetically conserved regions, nor do they need to be perfectly conserved across species.
Detection of clustered putative binding sites. Regulatory modules are often characterized by the presence of several binding sites for each of a small number of TFs (Howard and Davidson 2004). We identified regions of, at most, 2 kb that are significantly enriched in binding sites for one to up to five different TFs. To assign a ‘‘module score’’ to a given region, the five TFs with the highest total nonoverlapping scoring hits are chosen as tags for the putative module, and a P-value is assigned to the total score observed for the top one, two, three, four, or five tags. The number of tags for a given module is chosen so as to maximize the statistical significance of the hit density, so a short region that would be dense in sites for one TF would score well, as would a larger region with a few binding sites for each of a handful of factors. The P-value computation takes into consideration the number of factors involved (1–5), their total hit scores, the overall genome-wide frequency of their predicted hits, and the length and GC content of the region under evaluation (see Methods).
Our algorithm was used to scan the regions of the human genome that were alignable to the mouse and rat genome using the MULTIZ program (Blanchette et al. 2004; these regions cover 34% of the human genome). This resulted in the identification of 118,402 predicted modules, covering 2.88% of the human genome. Taken as a whole, this set of pCRMs, although likely to contain a non-negligible fraction of false positives, reveals a number of properties of human gene regions.
Although we considered putative modules of size up to 2000 bp, 58% of the pCRMs are less than 500 bp long, with an overall average length of 635 bp per CRM (see Supplemental Fig. S1A for a size histogram). This size distribution is quite close to that of the experimentally verified modules contained in the TRRD database (Kolchanov et al. 2002). However, we cannot exclude the possibility that some of the larger pCRMs are in fact made of more than one biological CRM. Modules have, on average, 3.1 tags (see Supplemental Fig. S1B), with shorter modules usually built from fewer tags than larger ones.
While the total number of individual sites predicted in phase (1) of our algorithm varies significantly from one PWM to another (see Supplemental Table S1), our procedure for correcting for low-specificity matrices ensures that no PWM is chosen as a tag too frequently. Supplemental Table S2 shows that tags are not seriously biased toward particular matrices, a sign that our algorithm for tag selection is sufficiently robust to avoid PWMs with low specificity. The PWM chosen as a tag the most often (5401 times, of 118,402 modules) is that for E2F, while the median PWM is selected as a tag in 704 modules. The PWMs that are the most often chosen for tags fall under two categories. The first is that of general promoter-associated factors, like E2F, ZF5, and TBP, which are indeed expected to bind a large number of regulatory regions. The second set of common tags consists of homeobox TFs (e.g., NKX family, POU family, etc.).
In silico validation of predicted modules
We evaluated the biological relevance of the pCRMs by measuring the extent to which they overlap known regulatory elements such as those compiled in the TRRD (Kolchanov et al. 2002), Transfac (Matys et al. 2003), and GALA (Giardine et al. 2003) databases. We also measured the overlap between the pCRMs and other putative regulatory elements, such as “promoter” regions (defined as the 1-kb region upstream of the transcription start sites [TSS] of all known genes), CpG islands (based on the UCSC Genome Browser annotation [Karolchik et al. 2003]), and DNaseI hypersensitive sites (Dorschner et al. 2004; Sabo et al. 2004;) from the Encode regions (Thomas et al. 2003). Figure 2A shows that despite the fact that only about 2.88% of the genome belongs to pCRMs, our predictions contain about 40% of the bases within modules annotated in GALA, 34% of the bases within Transfac binding sites, and 20% of the bases within the TRRD database. Our pCRMs are highly enriched within promoter regions, especially those containing CpG islands. Indeed, when considering the overlap between pCRMs and nonproximal (>1 kb upstream) annotated regulatory regions, our sensitivity (Fig. 2C) drops for all indicators except for the modules from the GALA database, though all remain severalfold higher than expected by chance (Fig. 2B,D). The significant enrichment for DNaseI hypersensitive sites is particularly interesting, as those represent an unbiased probing of chromatin structure. Although the function of these hypersensitive sites remains in most cases undetermined, many are likely to be CRMs.
Figure 2.
Sensitivity and enrichment of pCRMs for various regions of interest. (A) Sensitivity of the module predictions at varying score threshold, with respect to likely regulatory regions. Along the y-axis is the fraction of the bases within known regulatory regions that are predicted to belong to a pCRM. Along the x-axis is the number of predicted modules above a given threshold. Regions of interest are: 1 kb upstream: regions upstream of the TSS of Known Genes (based on the UCSC Genome Browser); Transfac sites: a set of 1209 experimentally verified binding sites from Transfac 7.2, mapped onto the human genome; TRRD modules: a set of 601 experimentally verified regulatory modules from the TRRD database; GALA modules: a set of 93 modules for the GALA database; CpG islands (based on the UCSC Genome Browser annotation); 1 kb upstream: regions upstream of the TSS of Known Genes that are not annotated as CpG islands; HS sites: a set of DNaseI hypersensitive sites from the Encode regions. (B) The fold enrichment is computed as the ratio between the size of the intersection between modules and regions of interest and the expected intersection size if modules were randomly positioned in the genome. (C,D) The analogous data, but restricting our attention to non proximal regulatory regions, i.e., those located more than 1 kb away from the TSS of the closest gene.
By definition, the sensitivity of our method for detecting annotated regulatory regions increases with the number of modules that are predicted. This increase is very rapid for the first ∼20,000 modules predicted, but the sensitivity for most indicators then increases more slowly. This observation is likely due to the fact that the modules that are the easiest to detect are those located in promoter regions. These also turn out to be the regions where most regulatory modules have been studied. However, the fact that our most reliable indicators of performance (TRRD modules, GALA modules, and, to a lesser extent, hypersensitive sites) continue to grow steadily after the first 20,000 pCRMs indicates that nonproximal modules can still be identified, and justifies considering a much larger set of modules.
Comparison to other genome-wide predictions
The ability of our algorithm to take advantage of interspecies TFBS conservation contributes in good part to the accuracy of the predictions. Indeed, the 34% of the human genome that lies within an alignment block with the mouse and rat genome contains 90% of bases within Transfac sites, 67% of those within TRRD modules, and 87% of those within GALA regulatory regions. Nonetheless, the sensitivity obtained by our pCRMs on these indicators remains three to five times higher than what would be obtained if modules were randomly predicted within the alignment blocks. To measure more accurately the extent to which sequence conservation alone can be used to predict known regulatory modules, sensitivity curves were computed based on the noncoding interspecies conserved regions identified by the PhastCons program (Siepel et al. 2005) (See Supplemental Fig. S2). The sensitivity of pCRMs is consistently 30%–70% higher than that of PhastCons elements for 1-kb “promoter” regions and TRRD and GALA modules, while it is comparable for Transfac and DNaseI hypersensitive sites. The advantage of pCRMs over PhastCons is most marked when only the highest-scoring half of each set of predictions is considered, in which case, the pCRMs sensitivity is at least twice that of PhastCons for all indicators.6 Overall, 41% of the bases within pCRMs lie within a PhastCons region (and 31% of PhastCons bases are within a pCRM), an 11-fold enrichment over what would be expected by chance.
Kolbe et al. (2004) and King et al. (2005) have developed a method called “regulatory potential,” which has been applied to the complete human genome to yield a set of CRM predictions. The method is trained to identify sequence features and interspecies conservation patterns that allow one to distinguish between a set of known regulatory regions and a set of nonfunctional regions. The overlap between the regulatory regions predicted by King et al. and our pCRMs is very significant—choosing a score threshold that results in about the same number of predicted bases as we get in our pCRMs (2.88% of the genome); more than 25% of the bases in pCRMs are also in King’s regions (nine times more than would be expected by chance). The accuracy of the two sets of predictions was compared based on the set of known regulatory regions used above, and none of the two methods appears significantly better than the other (see Supplemental Fig. S2), despite the fact that King’s method was trained on some of the specific regulatory regions used here for validation.
Experimental validation of predicted modules
In order to further validate our pCRMs, we took advantage of a technique called genome-wide location analysis (or ChIP-chip) (Ren et al. 2000; Iyer et al. 2001). This method allows for the large-scale identification of protein–DNA interactions as they occur in vivo. Briefly, proteins are cross-linked to DNA by treating live cells with formaldehyde and specific protein–DNA complexes are enriched by immunoprecipitation of fragmented chromatin using antibodies directed against a protein of interest. After reversal of the cross-links, the enriched DNA fragments are identified by hybridization onto DNA microarrays. We selected modules predicted to be bound by the estrogen receptor (ER), the E2F transcription factor 4 (E2F4), the signal transducer and activator of transcription 3 (STAT3), and the hypoxia-inducible factor 1 (HIF1) to print a DNA microarray. The microarray contains 758, 1370, 860, and 1882 modules predicted to be bound by ER, E2F4, STAT3, and HIF1, respectively. In the current study, the microarray was then probed by ChIP-chip for ER and E2F4 (see Methods for experimental details). After statistical analysis and experimental validation of the data (see Methods and Supplemental Table S3), we have identified 55 and 433 modules bound by ER and E2F4, respectively (see Supplemental Tables S4 and S5, respectively, and Table S6 for full ChIP-chip results). Approximately 3% of the 758 ER-predicted pCRMs on the microarray actually proved to be bound by ER, while 17% of the 1370 E2F4-predicted pCRMs on the microarray were bound by E2F4.
These numbers need to be considered as an underestimation of the actual specificity of the algorithm, since the protein–DNA interactions were tested in a single cell type, while TFs are known to regulate different sets of genes in different cell types, physiological conditions, and time in development (Zeitlinger et al. 2003; Hartman et al. 2005). For example, ER was tested in MCF-7, a breast cancer-derived cell line, due to its importance in breast cancer. ER, however, also plays important roles in many tissues such as ovaries, bone, brain, liver, and more. It is very likely that ER binds many pCRMs in some of these tissues, but not in MCF-7. In addition, the experiment was conducted under a single set of conditions (concentration of estradiol, time of treatment, etc.). For all of these reasons, it is difficult to determine the real accuracy of the algorithm.
Because our microarray contains predicted modules for four different TFs, the data can be used to assess the specificity of our TFBS predictions, e.g., to evaluate whether our prediction of which TFs should bind to each module is accurate. Among the 55 modules bound by ER, 44% (24/55, whereas 8/55 would be expected by chance) had indeed been selected for their ER-binding sites, and among the 433 modules bound by E2F4, 54% (236/433, whereas 147/433 would be expected by chance) had been selected for that factor. In addition to false-positive ChIP-chip signals or the failure of the algorithm to detect some binding sites, it is likely that binding of TFs through alternative mechanisms such as protein–protein interactions contributes to this result. For example, ER has been shown to be recruited to DNA by interaction with AHR to repress AHR-dependent gene regulation in an ER-responsive element-independent manner (Beischlag and Perdew 2005). It is important to note that our algorithm can only predict the binding of TF through direct DNA-binding interactions. It is likely that other TFs, in addition to those predicted here, may play roles in these modules. Of note, while 87% of the validated pCRMs for E2F4 were located in promoter regions, only 20% of those for ER were in these regions, confirming that our nonproximal pCRMs are also highly enriched for functional CRMs. Finally, Carroll et al. (2005) have used ChIP-chip on a tiling array to identify ER-binding sites on human chromosomes 21 and 22. Of the 57 regions they found to be bound by ER in MCF-7 cells, 14 overlap our predicted modules (five times more than expected by chance).
Despite the fact that the goal of this study is not to discuss specific interactions, we would like to highlight an interesting result that came out of the ChIP-chip experiments. While it is well known that the expression of the progesterone receptor gene PGR is up-regulated in breast cancer cells in response to estradiol, the absence of consensus estrogen response elements (ERE) in the two promoters driving its expression led to the suggestion that ER binds via other TFBSs (Petz et al. 2004). However, our data show that ER binds pCRMs present both ∼35 kb upstream of the TSS and ∼5 kb downstream of the 3′ end. Functional characterization of these pCRMs may reveal important clues about the molecular mechanisms implicated in long-range regulation by ER and other nuclear receptors (Carroll et al. 2005; Laganière et al. 2005).
A global view of the gene regulatory landscape
Having validated our predictions, we went on using them to study different global aspects of gene regulation. The genome-wide distribution of predicted modules is exemplified by Figure 3, which shows the pCRMs in a typical genomic region of human chromosome 11 containing the progesterone receptor gene PGR. The module density varies widely across the genome, with an average of four modules per 100 kb and a maximum of 44 modules per 100-kb window, covering from 0% to 55% of such a region. The presence of pCRMs is significantly correlated with the presence of a gene’s TSSs (correlation coefficient = 0.17, P-value < 10−308) on a local scale (10-kb window), but on a larger scale (1-Mb windows), no such correlation is observed. This indicates that the correlation between TSSs and pCRMs only extends to a few kilobases (Fig. 3B), and that distal pCRMs do not have strong location preferences relative to TSSs.
Figure 3.
Distribution of pCRMs along a region of chromosome 11. (A) A 10-megabase region from chromosome 11 is shown (coordinates 99, 308, 463–109, 308, 463). The position of the pCRMs (red) and the known genes (blue, from the UCSC Genome Browser) is shown. (B) A zoom in a 350-kilobase region containing the progesterone receptor gene (PGR) (coordinate 100, 400,000–100,750,000). The pCRM marked with an asterisk are those printed on our DNA microarray. (C) The composition of the Module M16589 is depicted as can be found in the PReMod database accompanying this study (http://genomequebec.mcgill.ca/PReMod). The position of the hits for five TRANSFAC matrices chosen as tags for this module is shown together with their individual scores.
As illustrated in Figure 3, some regions are rich in modules, but relatively poor in genes. In some cases, this could reflect the presence of many unknown protein-coding genes, or at least of many alternative TSSs. Another possible explanation is that some of these modules may be regulating the transcription of noncoding transcripts. Cumulating evidence indeed shows that much more transcription happens in the genome than what can be accounted for by traditional genes (Cawley et al. 2004; Cheng et al. 2005; The FANTOM Consortium 2005). Finally, this observation may be due to the presence of long-range enhancers, which may affect transcription of genes up to several hundreds of kilobases away (Bejerano et al. 2004; Baroukh et al. 2005; Woolfe et al. 2005). Clearly, a sizeable fraction of the module predictions is likely to be false positives, but there are no a priori reasons to expect false-positive predictions to cluster in any particular regions of the genome.
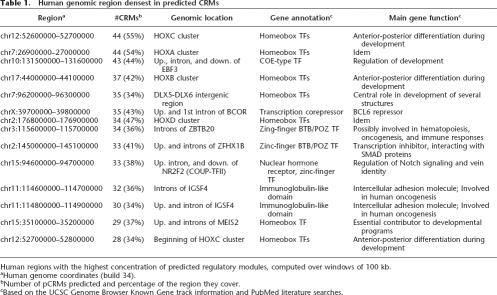
The genomic locations that are the densest in predicted modules (measured over 100-kb windows) are listed in Table 1. Most of these are located upstream, in the introns, or downstream of genes that are themselves TFs often involved in development. Among the 15 densest regions, we find parts of all four HOX clusters that operate differential genetic programs along the anterior–posterior axis of animal bodies (Alonso 2002), and regions near the EBF3, ZFHX1B, NR2F2, BCOR, MEIS2, and DLX5-6 genes, all of which are characterized TFs. The pCRMs in these regions have the unusual property of often being significantly conserved back to zebrafish and fugu, an indication that they may be part of the core regulatory mechanism of vertebrate development. There are 137 100-kb regions covered at least at 20% of CRMs, and these regions contain the TSSs of 115 genes with GO annotations (Harris et al. 2004). These genes are very strongly enriched for involvement in the regulation of transcription (79 genes, P-value 10−89), morphogenesis (24 genes, P-value 10−13), organogenesis (17 genes, P-value 3 × 10−5), and neurogenesis (10 genes, P-value 4 × 10−4), based on the Gostat program (Beissbarth and Speed 2004). We conjecture that genes involved in these processes often require very tight regulation, which in turn requires an elaborate set of regulatory modules. Notably, the presence in that group of ZBTB20, a poorly characterized gene encoding a predicted zinc finger TF, suggests the intriguing possibility that this TF may have a critical biological role, perhaps in regulating development.
Table 1.
Human genomic region densest in predicted CRMs
Human regions with the highest concentration of predicted regulatory modules, computed over windows of 100 kb.
aHuman genome coordinates (build 34).
bNumber of pCRMs predicted and percentage of the region they cover.
cBased on the UCSC Genome Browser Known Gene track information and PubMed literature searches.
There also exist regions that are very sparsely populated in predicted modules. One of the most striking examples is a 4-Mb region of chromosome 2 (chr2:123,000,001–127,000,000), of which <0.1% is covered by predicted modules. The region is somewhat of a gene desert, containing only one large gene annotated, hypothetical gene CNTNAP5. Other gene deserts are the opposite, quite rich in pCRMs. Many of those appear to be located in the vicinity of developmental TFs. For example, the homeobox gene MEIS1 is surrounded by a 1-Mb region devoid of any other TSS, but contains >130 kb of pCRMs.
Regulatory modules are preferentially located in specific regions relative to genes
We studied the position of pCRMs with respect to their closest gene. The genome was divided into several types of noncoding regions, i.e., upstream of a gene, 5′ UTR, 1st intron, internal introns, last intron, 3′ UTR, and downstream region. Within each type of region, we computed the fraction of bases included in a pCRM as a function of the distance to a reference point for each type of region (e.g., for upstream regions and 5′ UTR, the reference point is the TSS; see legend of Figure 4 for more details). This positional distribution was also compared with the positional distribution of a set of interspecies-conserved regions identified by the phastCons program (Siepel et al. 2005) on a set of aligned vertebrate genomes, using a conservation score threshold that results in a total number of noncoding bases predicted to be the same as the number of bases within pCRMs.
Figure 4.
Distribution of pCRMs relative to specific regions of genes. The genome was divided into several types of noncoding regions: upstream of a gene (dark blue), 5′ UTR (pink), 1st intron (yellow), internal introns (light blue), last intron (brown), 3′ UTR (aqua), and downstream region (dark blue). (A) For each type of region, the fraction of bases included in a pCRM is graphed as a function of the distance to a reference point. For upstream regions, 5′ UTR, and first intron the reference point is the gene’s TSS. For middle introns the closest 5′ or 3′ intron boundary is used. For the last intron, the 3′ UTR and the region 3′ of the last exon, the 3′ end of the mRNA is used. Note that the 3′ UTR is off the scale in A. (B) Same as in A, but different scales are used for the x- and y-axes in order to better show the characteristics of all regions.
From Figure 4, a number of striking observations are possible as follows:
Regions immediately surrounding TSSs are highly enriched for predicted modules. This was to be expected as this region often contains the promoter of the gene. More surprising is the presence of modules immediately downstream of the TSSs (either in the 5′ UTR or the first few kilobases of the first intron). These may represent alternative promoters for initiation downstream from the annotated transcripts. Alternatively, they may represent a yet underappreciated mode of activation that would take place from downstream proximal binding sites.
Regions surrounding the sites of termination of transcription are also highly enriched for modules. 3′ UTRs are essentially as enriched as 5′ UTRs for pCRMs, and module enrichment continues several kilobases past the end of the transcript, though to a lesser degree than in the upstream regions. At least two reasons may explain the presence of regulatory elements in the 3′ region of genes. First, these may represent enhancer type of regulatory elements that activate the upstream gene via a DNA-looping mechanism. Second, these may represent promoter elements driving noncoding transcript, antisense relative to the coding gene. Such antisense transcripts may regulate gene expression by a post-transcriptional mechanism (Cawley et al. 2004). Alternatively, these transcripts (or this transcription) may have biological roles of their own, independently of the coding transcript itself. For example, recent work in yeast showed that intergenic transcription could regulate gene expression by interfering with activation of a neighboring gene (Martens et al. 2004). It is possible that these TFBSs in the 3′ region of genes could give rise to antisense transcription that would interfere with sense transcription (Katayama et al. 2005). Recent analysis of the transcriptome of mammalian genomes revealed that a large proportion of all transcripts detected represent noncoding transcription (Kapranov et al. 2002; Cheng et al. 2005; The FANTOM Consortium 2005). Many of these noncoding transcripts map to the 3′ UTR of coding transcripts. ChIP-chip experiments performed on chromosome 21 and 22 (Cawley et al. 2004) have revealed that TFs can indeed bind these regions with a frequency higher than expected. These experimental data on chromosomes 21 and 22 are in agreement with our genome-wide predictions and likely reflect a yet understudied aspect of gene expression regulation.
Another surprising observation is that the density of modules is the lowest in regions located 10–50 kb upstream of the TSS and, symmetrically, 10–30 kb downstream of the end of transcription. This is unexpected, as one would expect that these regions (at least those upstream of the TSS) would be prime estate for transcriptional regulation. However, this is confirmed by the density of interspecies conserved elements, which is also at its lowest in those regions. We believe that this can be explained as follows: Thanks to their relative proximity to the TSS, regulatory elements in these regions may be allowed to contain fewer binding sites (or binding sites with less affinity), making them difficult to detect using our method. Alternatively, these regions may actually be depleted for regulatory elements. This could be due to constraints imposed by the chromatin structure of the nuclear architecture, making it more difficult for the DNA of these regions to come in physical proximity to the TSS. After the first 50 kb upstream of the TSS, the density of modules (and, to a lesser extent, of conserved regions) starts increasing with the distance to the TSS, with regions located >200 kb upstream of the TSS, being about 50% more densely populated in modules than the –50 to –10 kb region. We believe that this may be explained by the fact that regulatory modules that are located very far from the gene they regulate would often require many strong binding sites, making their computational detection easier. The symmetric effect is observed in regions downstream of genes, although at these large distances it is unclear whether these modules would regulate the sense or antisense transcription.
The density of predicted modules in intronic regions is very low in the close vicinity of exons (except the first and last ones), but increases with the distance to the closest exon. Although some of the intronic pCRMs may turn out to be splicing regulatory regions, this is unlikely to be the case for a large fraction of them, as intronic splicing elements usually cluster near exon boundaries (Sorek and Ast 2003). Instead, we speculate that CRMs within these very large introns may be located in genes that require tighter transcriptional regulation, resulting in a higher module density in these regions.
Although the module density usually follows closely the interspecies conservation density, a few notable exceptions indicate that our module predictions are doing more than merely detecting conserved regions. First, the regions surrounding the TSS (on either side) are much richer in modules than in conserved regions. Second, the 1-kb regions immediately flanking internal exons tend to be highly conserved (Sorek and Ast 2003) and they are believed to be involved in splicing regulation. However, these regions are depleted from pCRMs, as indeed these regions are not involved in transcriptional regulation and lack the signature sought by our algorithm. As a side note, pCRMs are also twofold depleted in known RNA genes, although these too tend to be well conserved evolutionarily.
Specific TFs target different regions relative to their target genes
As described above, our predictions, when taken altogether, are enriched in the 5′ and 3′ region of known genes. When broken down into predictions for individual TFs, however, a great variability in observed. For example, our predictions of ER modules (e.g., modules predicted to contain at least one high-scoring ER-binding site) are enriched in regions located more than 10 kb upstream of known genes, while our predictions for E2F4 are enriched in the proximal 5′ region of known genes. This suggests that ER functions mainly through distal, enhancer-like elements, while E2F4 regulates gene transcription via promoter-proximal elements. Notably, evidence in the literature supports this hypothesis (see Blais and Dynlacht 2005; Carroll et al. 2005). Importantly, our ChIP-chip data also supports this model. Indeed, despite the fact that pCRMs printed on the array were uniformly distributed with respect to genes, only 20% of the pCRMs bound by ER in our ChIP-chip experiments were within 1 kb on either side of the TSS, while the proportion is of 87% for the pCRM bound by E2F4. Based on this observation, we have computed the location preferences of each of the 229 TF families represented by the PWMs used in our predictions (see Figure 5 and Supplemental Table S7). Figure 5 shows that more than 70 of the 229 TFs families considered exhibit a significant enrichment for one or more types of genomic regions (see Methods). These TFs separate quite clearly into two groups with very little overlap. A number of TFs show preference for distal positions, mostly those located more than 100 kb upstream of the TSS, and are also often enriched within introns. This set of TFs is enriched for factors containing homeo domains or basic helix-loop-helix domains and are often involved in regulating development. Some of these factors have indeed been shown to bind distal modules and activate transcription during early development (Bejerano et al. 2004; Woolfe et al. 2005). Notably, we find no TFs enriched for introns only (except within 1 kb downstream of the TSS), which indicates that regulatory modules located in introns are of the same type as those located far away from genes. In fact, it is likely that certain intronic modules do not regulate the gene in which they are located, but rather another gene located nearby, as reported recently for sonic hedgehog (Sagai et al. 2005)
Figure 5.
Many TFs preferentially bind to specific regions relative to the TSS of their target genes. A heat map of the enrichment (represented as a Z-score) of a TF for different regions relative to TSSs is shown. Regions in red are highly enriched for binding sites for the given TF, while those in blue are depleted. The regions shown on the x-axis are as follows: >100kb upstream, pCRMs located more than 100 kb upstream from a TSS; 10–100kb upstream, pCRMs located >10 kb, but <100 kb upstream from a TSS; 1–10kb upstream, pCRMs located >1 kb but <10 kb upstream from a TSS; 0–1kb upstream, pCRMs located within 1 kb upstream of a TSS; 1kb 1st intron, intronic pCRMs located within 1 kb downstream of the TSS of a gene; 10kb 1st intron, intronic pCRMs located within 10 kb downstream of a TSS; intron, intronic pCRM located >10 kb from the TSS; 0–1kb down, pCRM located within 1 kb from the 3′ end of a gene; 1–10kb down, pCRM located >1 kb but <10 kb downstream from the 3′ end of a gene. See Methods for details on the computation of Z-scores.
A second set of TFs preferentially binds within 1 kb of the TSSs. This set is enriched for leucine zipper TF and factors from the Ets family. Notably, most of these factors, contrary to what is observed for those binding distal sites, are involved in basic cellular functions. Among the best-known examples we found NF-Y, E2F, CREB, ATF, and others. Interestingly, and much to our surprise, most of these TFs show a clear preference for either the 1 kb upstream or the 1 kb downstream of the TSS, but not both. The most striking example is Nuclear Factor Y (NF-Y), which is highly enriched 1 kb upstream, but highly depleted 1 kb downstream of the TSS. This preference may reflect a mechanistic characteristic of these TFs. Finally, note that when we computed enrichment statistics based on all genome-wide predicted TFBSs instead of based only on those located in modules, much fewer TFs obtained significant enrichment in any given type of region, indicating that our pCRMs are effective at reducing the false-positive rate in TFBS predictions.
Long-range correlation of TFBS predictions
We observe that the closer together two modules are on the genome, the more likely they are to contain predicted binding sites for the same factors. Part of this is simply due to isochors, those broad variations of GC content along the genome (International Human Genome Sequencing Consortium 2001). However, even after correcting for this factor (see Methods), a number of TFs show significant long-range correlation between their predicted sites (Supplemental Fig. S3; Supplemental Table S8). This is likely to be due to the fact that if several regulatory modules regulate a gene, they are likely to be bound by a similar set of TFs. Not surprisingly, most of the TFs that exhibit long-range correlation are those that show preferences for binding sites located more than 10 kb upstream of the TSS. The set of nearby pCRMs that contain binding sites for similar TFs tends to be located in large intergenic or intronic regions and they tend to be located near genes encoding TFs.
Predicted TFBSs induce correlated tissue-specific gene expression
Comparison of TF-binding data with gene expression data in yeast showed that genes bound by a common set of TFs tend to be coregulated (Lee et al. 2002). Such a correlation is expected to occur in mammalian cells as well, but was never thoroughly tested because of the lack of genome-wide data for TF binding. Our predicted module data allows us to investigate this question. For each TF family in our study, a set of putatively regulated genes was identified as those with at least one predicted high-scoring site in a pCRM located within 10 kb upstream of the TSS. We computed the average pairwise Pearson correlation coefficient between tissue-specific expression levels of the genes of the set using expression data from 79 human cell types or tissues from the GNF Atlas 2 (Su et al. 2004). A total of 27 of the 229 TF families are associated to a significant expression correlation (P-value < 0.01, false-discovery rate (FDR) = 8%; see Supplemental Table S9). We repeated our correlation analysis, this time measuring the expression correlation for genes sharing binding sites for pairs of TFs. Of the 26,106 pairs of TF families considered, 595 are associated to a significant expression correlation (P-value < 0.01, FDR = 43%) (See Supplemental Table S10 for a complete list). For example, most of the 20 genes that have a pCRM containing OCT-1 and BACH1-binding sites are highly expressed in various brain tissues, excluding the cerebellum and the olfactory bulb, and in the pituary gland. While the role of OCT-1 in brain cells has already been characterized (Givens et al. 2004), its association with BACH1 has not been reported before.
Since most TFs are only expressed in a subset of the 79 cell types considered, they are unlikely to induce significant coexpression when measured over all 79 cell types. In order to identify transcription factors regulating expression in specific cell types, we analyzed each pair of TF and cell type. For each pair, the average expression level of the genes associated with predicted binding sites for the TF was computed and its significance assessed by a permutation test. Of the 229 × 79 = 18,091 possible (TF-cell type) pairs, we found 119 where genes are overexpressed (P-value < 0.001, FDR = 15%), and 78 where genes are underexpressed (P-value < 0.001, FDR = 23%). Table 2 lists the pairs with the most significant associations (see Supplemental Table S11 for the complete list). For example, the genes associated with pCRMs for MyoD tend to be highly expressed in skeletal muscle cells, while those associated to Ets are highly expressed in white blood cells. Both the role of MyoD in skeletal muscles and that of Ets in blood cells are very well characterized, thereby validating the approach.
Table 2.
Tissue-specific expression for genes predicted to be regulated by various types of transcription factors
aTissues expressing high level of putative target for the given TF.
bTissues expressing low level of putative target genes for the given TF. See Methods for details.
We also discovered associations that are not well characterized. For instance, we found that genes around pCRMs for NF-Y tend to have low expression in the ciliary and superior cervical ganglia and high expression in thymus and lymphoblasts. NF-Y binds an element called the CCAAT box, which has been reported to be present within promoters of genes activated during peptide presentation in antigen presenting cells (APC) (Mach et al. 1996) and within the promoters of housekeeping genes such as those regulated during the cell cycle (Mantovani 1999). From this literature, one would not have predicted a role for NF-Y in the brain and the thymus, but the fact that ciliary and ganglia cells are not (or only slowly) dividing and that some APC originate from thymus (Choi et al. 2005) is however consistent with our findings.
The average expression levels were also computed for the set of genes associated with each pair of TFs. Of the roughly 2 million triplets (TF1, TF2, cell-type) tested, 5242 triplets show significant overexpression (P-value < 0.001, FDR <39%), while 6407 triplets show significant underexpression (P-value < 0.001, FDR <31%; see Supplemental Table S12).
A searchable public database of predicted regulatory modules
The modules predicted by the algorithm were stored in a database with a Web-based interface (http://genomequebec.mcgill. ca/PReMod). The database supports a variety of queries and contains hyperlinks pointing to the NCBI Entrez of the closest gene. The module information includes its genomic position as well as its TFBS content. A graphical view of the TFBS distribution of the highest scoring matrices is also provided (see, for example, Fig. 3C). Queries can reveal relationships such as the set of modules associated with a specific matrix, the set of modules located in the vicinity of a gene of interest, the set of the modules located within a specific distance from any gene, the set of modules associated with CpG islands, etc. Output from queries can be viewed as html or Excel tables. Genomic sequence of the whole set of modules can also be downloaded in fasta format from the Web site.
Conclusions
Using the literature as a guideline, we have identified a set of rules describing the architecture of DNA regulatory elements and used them to build an algorithm allowing us to explore the regulatory potential of the human genome. Although the error rate in CRM predictions is likely to be relatively high, the statistical power obtained through a large-scale, genome-wide approach revealed new insights into the biology of transcriptional regulation. Among other things, we observe a strong enrichment for pCRMs in regions at the 3′ end of genes. By concentrating on predicted TF-binding sites within pCRMs, we are able to improve the specificity of individual TFBS predictions, which allows the detection of signals that could not be seen otherwise. For example, we noted that a significant number of TFs have a strong bias for regulating genes either from a great distance or from promoter-proximal binding sites. Noteworthy is the fact that most TFs that preferentially work from a large distance are involved in development, while those predicted to work from promoter-proximal sites tend to regulate genes involved in basic cellular processes. We have identified a set a TFs that are predicted to play important roles in specific tissues, including cells and tissues issued from tumors and metastases. Finally, our data provides a starting point for the elaboration of human gene networks.
In a bootstrap-like fashion, several of the features derived from our pCRMs could be used to design improved CRM prediction algorithms. For example, the fact that specific TFs prefer binding at specific locations with respect to genes and that CRMs tend to organize in larger and looser clusters often containing binding sites for similar sets of factors could allow improved predictions.
We expect that the database containing the modules predicted in this study may speed up the discovery and experimental validation of CRMs. Finally, deeper data-mining approaches are likely to yield a plethora of specific testable biological hypotheses.
Methods
Transfac position weight matrices
A set of 481 vertebrate PWMs from Transfac 7.2 (Matys et al. 2003) was used for the analysis. Pseudocounts were introduced to regularize matrices based on few known sites (Durbin et al. 1998). Many PWMs represent the same or very similar factors. This does not cause any problem to our CRM prediction algorithm (since it excludes overlapping sites), but it is undesirable for downstream analyses of individual TF properties, e.g., localization with respect to the genes and tissue-specific expression. For these sections of the study, PWMs were grouped into 229 families based on the following rule: If many related TFs had individual PWMs, but Transfac also contained a generic PWM for the family, then only that generic matrix was used.
Module prediction algorithm
The outline of our module prediction algorithm is provided in Figure 1. We used a genome-wide multiple alignment of the human, mouse, and rat genomes (versions hg16, mm3, and rn2) produced by the MULTIZ program (Blanchette et al. 2004) and available from the UCSC Genome Browser (Karolchik et al. 2003). Only regions within MULTIZ alignment blocks are considered in what follows. These regions cover 34% of the human genome. For each of the 481 PWMs, individual binding sites are first predicted as follows. The human, mouse, and rat genomic regions are first scanned separately, on both strands, and a log-likelihood ratio score is computed in the standard way (Durbin et al. 1998). The only improvement is that we use a set of 3rd-order Markov models for background, and the choice of model depends on the local GC-content of the 1-kb region surrounding the position. Twenty different Markov models have been trained, based on nonrepetitive, noncoding human genomic regions with 0%–5% GC, 5%–10% GC, 95%–100% GC, and at every position the most appropriate background model is used.
Species-specific scores are then mapped onto the alignment and for each alignment column p and PWM m, we compute: hitScorealn(m,p) = hitScoreHum(m,p) + 1/2 max(0, hitScoreMou(m,p) + hitScoreRat(m,p)). Thus, hitScorealn(m,p) will be high if all three species have a high-scoring site at position p. Notice that if the hit score of human is very high, the resulting hitScorealn may be relatively good even if mouse and/or rat do not have high-scoring hits at that position. This allows us to predict human-specific binding sites, provided that they are very good matches to the PWM considered. Once the alignment scan is completed, only positions with hitScorealn(m,p) > 10 are retained to construct modules. This results in a total number of predicted sites that varies from 1.5 million for E2F (M00103) to about 8000 for Hogness (M00316), many of which are expected to be false positives (see Supplemental Table S1).
We now discuss how to compute moduleScore(p1..p2) for the alignment region going from position p1 to p2 of human. We first define TotalScore(m, p1..p2) to be the sum of the hitScoresaln of all nonoverlapping hits for m in the region p1..p2. Formally, letting Hm be the set of all hits for matrix m in region p1..p2, we have TotalScore(m,p1..p2) = max {H ⊆ Hm s.t. hits in H do not overlap} Σh ∈ H hitScore(m,p).
The optimization problem of choosing the best set of nonoverlapping hits is solved heuristically, using a greedy algorithm that iteratively selects the hit with the maximal score that does not overlap with the other hits previously chosen. For each matrix and each region, a P-value is assigned to the TotalScore observed, measuring the probability that a random region of the human–mouse–rat alignment would have a total score that would exceed the observed one. This P-value takes into consideration the length and GC-content of the region considered, as well as the overall frequency and score distribution of hits predicted for that matrix in the genome. This allows for a region dense in hits for a rare matrix (i.e., one with few hits in the genome) to obtain a higher score than a region equally dense in hits for a more common matrix. Matrices that tend to have a large number of hits throughout the genome are thus penalized. More precisely, for each matrix m, GC-content g and window length l, the distribution of TotalScore is estimated empirically through simulation, repeating 10 million times the following procedure: (1) choose l random positions from alignment regions with GC-content g and (2) compute the TotalScore of the set of positions selected, assuming that the l positions chosen form a contiguous region.
The score of a candidate module is computed based on one to five PWMs called tags. The first tag for region p1..p2 is the matrix with the most significant TotalScore, i.e., tag1 = argminm∈PWMs pValue(TotalScore(m,p1..p2)). The regions belonging to the hits selected for tag1 are then masked out and the TotalScores for each matrix are recomputed, excluding hits overlapping those of tag1. The second tag is then the matrix that achieves the most significant totalScore, and its occurrences are masked out. The process is repeated until five tags are selected, if possible. Finally, we define moduleScore(p1..p2) = max{k = 1..5} –log (pValueMaxUnif (k, 481, ∏i =1..k pValue(totalScore(tagk, p1..p2)))), where pValueMaxUnif(k, 481, a) is the probability that the product of k random variables, each defined as the maximum of 481 uniform(0,1) random variables, is smaller than a.7 A module can thus consist of one to five tags, depending on which number of tags yields the highest statistical significance.
The above procedure was used to search for modules of maximal length 100, 200, 500, 1000, and 2000bp.8 For each window size, regions with moduleScore > 10 (i.e., P-value < e−10) were identified. This choice of threshold is somewhat arbitrary, but results in a total number of bases predicted in pCRMs to be ∼2.88% of the genome, a reasonable upper bound for the fractions of bases in regulatory regions. To address the fact that many of these modules overlap each other, a greedy algorithm was used to repeatedly select the highest-scoring module not overlapping any of the previously selected higher-scoring modules. This resulted in the set of 118,402 nonoverlapping modules studied in this work. Predictions were then mapped onto the latest human assembly (hg17) using the liftOver program (Karolchik et al. 2003; <0.1% of modules could not be mapped onto the new assembly and were discarded).
Microarray design and production
A subset of the pCRMs was selected to build a microarray to be used for ChIP-chip validation experiments. For each TFs among ER, HIF1, STAT3, and E2F4, at most 50 pCRMs were randomly selected for each combination of the following categories: (1) module score: High vs. non-high; (2) totalScore for the given TF: High vs. non-high; (3) genomic location with respect to closest TSS: 10–100 kb upstream, 800 bp–10 kb upstream, -800 to +200 bp, +200 bp to +1000 bp, +1 kb to +10 kb, 0–10 kb downstream of 3′ UTR, or other. Most combinations could be not filled up to their quota. Each pCRM selected was extended symmetrically to a size of 1 kb, excluding repetitive regions. Primer pairs were designed for each region, using the Primer3 algorithm (Rozen and Skaletsky 2000), and the specificity was tested in silico by using a virtual PCR algorithm (Lexa et al. 2001). When the primer pair gave no satisfactory virtual PCR results, a new primer pair was designed by using Primer3 and tested again. The process was iterated three times to generate primer pairs predicted to be efficient to amplify regions from human genomic DNA for almost all of our selected pCRMs. This primer design pipeline allowed us to design primer pairs to amplify pCRMs from human genomic DNA with a success rate of ≈85%.
ChIP-chip assay and data analysis
ER ChIP-chip experiments were performed as described previously (Laganière et al. 2005). E2F4 ChIP-chip experiments were performed as follows: T98G cells (ATCG) were grown in DMEM containing 10% FBS and arrested through contact inhibition by allowing cells to reach confluence. Medium was changed after the second day of confluence and cells harvested on the third day. Confluent T98G cells were fixed with 1% formaldehyde, rinsed twice with PBS, and harvested. The cell pellet was lysed and sonicated to obtain DNA fragments of 600 bp on average. ChIP was performed using anti-E2F4 antibody (sc-1082, Santa-Cruz) and Dynabeads (Dyna). ChIP samples and nonimmunoprecipitated fragments were blunted with T4 DNA polymerase and ligated to unidirectional linkers. The DNA was then amplified by LM–PCR and labeling carried out post PCR by incorporation of Cy5 or Cy3-dUTP using Klenow polymerase reaction. Detail protocol can be found at http://www.ircm.qc.ca/microsites/francoisrobert/en.
Data were normalized and triplicates were combined using a weighted average method as described previously (Ren et al. 2000). The P-value threshold used for the analysis was established by testing the enrichment of 10 targets for each of the following P-value intervals for both ER and E2F4 ChIPs using quantitative PCR with SYBR Green: <0.001, 0.001–0.005, 0.005–0.01, 0.01–0.05, 0.05–0.1, 0.1–0.5, 0.5–1. The results of this validation process are shown in Supplemental Table S1. Using P < 0.01 (ER) and P < 0.1 (E2F4), virtually all targets are bona fide binding sites (see Supplemental Tables S2 and S3). All microarray data will be deposited to ArrayExpress.
Statistical significance of TF location preferences and spatial correlation
We used a permutation test to estimate the statistical significance of the observed number of binding sites predicted in each type of region of the genome. Given the set of all predicted sites for all TFs, we first removed from consideration all but one of the hits of a TF within a given module. Each module thus contains at most one binding site for a given TF. To perform our permutation test, we repeatedly randomly chose two sites for two different factors, and exchanged their labels (but kept the original positions), provided they both lie in regions of the same GC-content (within 1% difference, measured over 1 kb). The scrambling procedure was sufficiently repeated often to reach a random distribution, at which point the number of sites in each region was counted. The experiment was repeated 100 times, from which the expectation and variance of the count of each TF in each region was estimated and the Z-score calculated. Notice that this procedure preserves the varying density of binding sites across the genome (since only labels, but not positions, are modified), as well as the local GC-content preferences of each TF. To estimate the significance of the long-range spatial correlations observed between sites of a given TF, a similar permutation test was applied and the observed number of co-occurrence within a given distance was compared with those obtained in the permuted data sets, allowing to compute a Z-score for each TF and distance interval.
Correlation between predicted TFBS and tissue-specific gene expression
For each TF, a set of putative target genes was defined as the genes with at least one high-scoring predicted site for that TF within a pCRM and within 10 kb of the TSS. The average expression level of these genes in each of 79 tissues (GNF Atlas II) was calculated and its significance was estimated using a permutation test. Tissues showing overexpression or underexpression with Z-score > 5 are reported in Table 2.
Acknowledgments
This work was funded by grants from Génome Québec and Génome Canada (M.B., V.G., B.C., and F.R.) and by the Canadian Institutes for Health Research (V.G). A.R.B. is a recipient of a doctoral fellowship from the IRCM/CIHR Cancer Research Program. X.C. is a recipient of a Génome Québec Comparative and Integrative Genomics Program. J.L. is a recipient of a U.S. Department of Defense Breast Cancer Research Program Predoctoral Traineeship Award (#W8IWXH-04-1-0399). F.R. holds a new investigator award from the CIHR. We thank Adam Siepel for his PhastCons data, UCSC Genome Browser group for their support, and John Stamatoyannopoulos for the DNaseI hypersensitive regions data.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.4866006
Since PhastCons was designed to detect any type of region under selective pressure, many of its noncoding predictions are likely to have other nonregulatory functions.
Note that the formula for moduleScore is actually an approximation of the true P-value, for the following reasons: (1) Since competition for space between different tags is not modeled, the computed P-value of the total score of the 2nd, 3rd, 4th, and 5th tags are slightly conservative; (2) since the totalScores are discrete variables (but with a very large number of possible values), the approximation with a continuous uniform distribution introduces a small error; (3) since the moduleScore is obtained by selecting the best of five P-values, a multiple hypothesis testing correction should be applied. However, since we are mostly interested in the ranking of modules, this correction would make no difference.
Only a small number of maximal lengths could be tried, as the calculation of the TotalScore P-values are computationally expensive and depend on that length.
References
- Aerts S., Loo P.V., Thijs G., Moreau Y., Moor B.D., Loo P.V., Thijs G., Moreau Y., Moor B.D., Thijs G., Moreau Y., Moor B.D., Moreau Y., Moor B.D., Moor B.D.2003Computational detection of cis-regulatory modules. Bioinformatics (Suppl 2) 19II5–II14. [DOI] [PubMed] [Google Scholar]
- Aerts S., Loo P.V., Moreau Y., Moor B.D., Loo P.V., Moreau Y., Moor B.D., Moreau Y., Moor B.D., Moor B.D. A genetic algorithm for the detection of new cis-regulatory modules in sets of coregulated genes. Bioinformatics. 2004;20:1974–1976. doi: 10.1093/bioinformatics/bth179. [DOI] [PubMed] [Google Scholar]
- Alkema W.B.L., Johansson O., Lagergren J., Wasserman W.W., Johansson O., Lagergren J., Wasserman W.W., Lagergren J., Wasserman W.W., Wasserman W.W. MSCAN: Identification of functional clusters of transcription factor binding sites. Nucleic Acids Res. 2004;32:W195–W198. doi: 10.1093/nar/gkh387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso C.R. Hox proteins: Sculpting body parts by activating localized cell death. Curr. Biol. 2002;12:R776–R778. doi: 10.1016/s0960-9822(02)01291-5. [DOI] [PubMed] [Google Scholar]
- Bailey T.L., Noble W.S., Noble W.S.2003Searching for statistically significant regulatory modules. Bioinformatics 19 : II16–II25. [DOI] [PubMed] [Google Scholar]
- Bajic V.B., Choudhary V., Hock C.K., Choudhary V., Hock C.K., Hock C.K. Content analysis of the core promoter region of human genes. In Silico Biol. 2004;4:109–125. [PubMed] [Google Scholar]
- Baroukh N., Ahituv N., Chang J., Shoukry M., Afzal V., Rubin E.M., Pennacchio L.A., Ahituv N., Chang J., Shoukry M., Afzal V., Rubin E.M., Pennacchio L.A., Chang J., Shoukry M., Afzal V., Rubin E.M., Pennacchio L.A., Shoukry M., Afzal V., Rubin E.M., Pennacchio L.A., Afzal V., Rubin E.M., Pennacchio L.A., Rubin E.M., Pennacchio L.A., Pennacchio L.A. Comparative genomic analysis reveals a distant liver enhancer upstream of the COUP-TFII gene. Mamm. Genome. 2005;16:91–95. doi: 10.1007/s00335-004-2442-9. [DOI] [PubMed] [Google Scholar]
- Beischlag T.V., Perdew G.H., Perdew G.H. ER α-AHR-ARNT protein-protein interactions mediate estradiol-dependent transrepression of dioxin-inducible gene transcription. J. Biol. Chem. 2005;280:21607–21611. doi: 10.1074/jbc.C500090200. [DOI] [PubMed] [Google Scholar]
- Beissbarth T., Speed T.P., Speed T.P. GOstat: Find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics. 2004;20:1464–1465. doi: 10.1093/bioinformatics/bth088. [DOI] [PubMed] [Google Scholar]
- Bejerano G., Pheasant M., Makunin I., Stephen S., Kent J., Mattick J., Haussler D., Pheasant M., Makunin I., Stephen S., Kent J., Mattick J., Haussler D., Makunin I., Stephen S., Kent J., Mattick J., Haussler D., Stephen S., Kent J., Mattick J., Haussler D., Kent J., Mattick J., Haussler D., Mattick J., Haussler D., Haussler D. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- Blais A., Dynlacht B.D., Dynlacht B.D. Constructing transcriptional regulatory networks. Genes & Dev. 2005;19:1499–1511. doi: 10.1101/gad.1325605. [DOI] [PubMed] [Google Scholar]
- Blanchette M., Kent W.J., Riemer C., Elnitski L., Smit A.F.A., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Kent W.J., Riemer C., Elnitski L., Smit A.F.A., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Riemer C., Elnitski L., Smit A.F.A., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Elnitski L., Smit A.F.A., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Smit A.F.A., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Rosenbloom K., Clawson H., Green E.D., Clawson H., Green E.D., Green E.D., et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulyk M.L. Computational prediction of transcription-factor binding site locations. Genome Biol. 2003;5:201. doi: 10.1186/gb-2003-5-1-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll J.S., Liu X.S., Brodsky A.S., Li W., Meyer C.A., Szary A.J., Eeckhoute J., Shao W., Hestermann E.V., Geistlinger T.R., Liu X.S., Brodsky A.S., Li W., Meyer C.A., Szary A.J., Eeckhoute J., Shao W., Hestermann E.V., Geistlinger T.R., Brodsky A.S., Li W., Meyer C.A., Szary A.J., Eeckhoute J., Shao W., Hestermann E.V., Geistlinger T.R., Li W., Meyer C.A., Szary A.J., Eeckhoute J., Shao W., Hestermann E.V., Geistlinger T.R., Meyer C.A., Szary A.J., Eeckhoute J., Shao W., Hestermann E.V., Geistlinger T.R., Szary A.J., Eeckhoute J., Shao W., Hestermann E.V., Geistlinger T.R., Eeckhoute J., Shao W., Hestermann E.V., Geistlinger T.R., Shao W., Hestermann E.V., Geistlinger T.R., Hestermann E.V., Geistlinger T.R., Geistlinger T.R., et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Cawley S., Bekiranov S., Ng H.H., Kapranov P., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Bekiranov S., Ng H.H., Kapranov P., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Ng H.H., Kapranov P., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Kapranov P., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Sementchenko V., Cheng J., Williams A.J., Cheng J., Williams A.J., Williams A.J., et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- Cheng J., Kapranov P., Drenkow J., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Kapranov P., Drenkow J., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Drenkow J., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Patel S., Long J., Stern D., Tammana H., Helt G., Long J., Stern D., Tammana H., Helt G., Stern D., Tammana H., Helt G., Tammana H., Helt G., Helt G., et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- Choi E.Y., Jung K.C., Park H.J., Chung D.H., Song J.S., Yang S.D., Simpson E., Park S.H., Jung K.C., Park H.J., Chung D.H., Song J.S., Yang S.D., Simpson E., Park S.H., Park H.J., Chung D.H., Song J.S., Yang S.D., Simpson E., Park S.H., Chung D.H., Song J.S., Yang S.D., Simpson E., Park S.H., Song J.S., Yang S.D., Simpson E., Park S.H., Yang S.D., Simpson E., Park S.H., Simpson E., Park S.H., Park S.H. Thymocyte-thymocyte interaction for the efficient positive selection and maturation of CD4 T cells. Immunity. 2005;23:387–396. doi: 10.1016/j.immuni.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Davidson E. Genomic regulatory systems: Development and evolution, Academic Press; NY.: 2001. [Google Scholar]
- Dorschner M.O., Hawrylycz M., Humbert R., Wallace J.C., Shafer A., Kawamoto J., Mack J., Hall R., Goldy J., Sabo P.J., Hawrylycz M., Humbert R., Wallace J.C., Shafer A., Kawamoto J., Mack J., Hall R., Goldy J., Sabo P.J., Humbert R., Wallace J.C., Shafer A., Kawamoto J., Mack J., Hall R., Goldy J., Sabo P.J., Wallace J.C., Shafer A., Kawamoto J., Mack J., Hall R., Goldy J., Sabo P.J., Shafer A., Kawamoto J., Mack J., Hall R., Goldy J., Sabo P.J., Kawamoto J., Mack J., Hall R., Goldy J., Sabo P.J., Mack J., Hall R., Goldy J., Sabo P.J., Hall R., Goldy J., Sabo P.J., Goldy J., Sabo P.J., Sabo P.J., et al. High-throughput localization of functional elements by quantitative chromatin profiling. Nat. Methods. 2004;1:219–225. doi: 10.1038/nmeth721. [DOI] [PubMed] [Google Scholar]
- Durbin R., Eddy S., Krogh A., Mitchison G., Eddy S., Krogh A., Mitchison G., Krogh A., Mitchison G., Mitchison G.1998. , Biological sequence analysis. Cambridge University Press; Cambridge, UK. [Google Scholar]
- The FANTOM Consortium The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Frith M.C., Li M.C., Weng Z., Li M.C., Weng Z., Weng Z. Cluster-Buster: Finding dense clusters of motifs in DNA sequences. Nucleic Acids Res. 2003;31:3666–3668. doi: 10.1093/nar/gkg540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardine B., Elnitski L., Riemer C., Makalowska I., Schwartz S., Miller W., Hardison R.C., Elnitski L., Riemer C., Makalowska I., Schwartz S., Miller W., Hardison R.C., Riemer C., Makalowska I., Schwartz S., Miller W., Hardison R.C., Makalowska I., Schwartz S., Miller W., Hardison R.C., Schwartz S., Miller W., Hardison R.C., Miller W., Hardison R.C., Hardison R.C. GALA, a database for genomic sequence alignments and annotations. Genome Res. 2003;13:732–741. doi: 10.1101/gr.603103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Givens M.L., Kurotani R., Rave-Harel N., Miller N.L., Mellow P.L., Kurotani R., Rave-Harel N., Miller N.L., Mellow P.L., Rave-Harel N., Miller N.L., Mellow P.L., Miller N.L., Mellow P.L., Mellow P.L. Phylogenetic footprinting reveals evolutionarily conserved regions of the gonadotropin-releasing hormone gene that enhance cell-specific expression. Mol. Endocrinol. 2004;18:2950–2966. doi: 10.1210/me.2003-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M., Liu J.S., Liu J.S. De novo cis-regulatory module elicitation for eukaryotic genomes. Proc. Natl. Acad. Sci. 2005;102:7079–7084. doi: 10.1073/pnas.0408743102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris M.A., Clark J., Ireland A., Lomax J., Ashburner M., Foulger R., Eilbeck K., Lewis S., Marshall B., Mungall C., Clark J., Ireland A., Lomax J., Ashburner M., Foulger R., Eilbeck K., Lewis S., Marshall B., Mungall C., Ireland A., Lomax J., Ashburner M., Foulger R., Eilbeck K., Lewis S., Marshall B., Mungall C., Lomax J., Ashburner M., Foulger R., Eilbeck K., Lewis S., Marshall B., Mungall C., Ashburner M., Foulger R., Eilbeck K., Lewis S., Marshall B., Mungall C., Foulger R., Eilbeck K., Lewis S., Marshall B., Mungall C., Eilbeck K., Lewis S., Marshall B., Mungall C., Lewis S., Marshall B., Mungall C., Marshall B., Mungall C., Mungall C., et al. The Gene Ontology GO database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman S.E., Bertone P., Nath A.K., Royce T.E., Gerstein M., Weissman S., Snyder M., Bertone P., Nath A.K., Royce T.E., Gerstein M., Weissman S., Snyder M., Nath A.K., Royce T.E., Gerstein M., Weissman S., Snyder M., Royce T.E., Gerstein M., Weissman S., Snyder M., Gerstein M., Weissman S., Snyder M., Weissman S., Snyder M., Snyder M. Global changes in STAT target selection and transcription regulation upon interferon treatments. Genes & Dev. 2005;19:2953–2968. doi: 10.1101/gad.1371305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard M.L., Davidson E.H., Davidson E.H. cis-Regulatory control circuits in development. Dev. Biol. 2004;271:109–118. doi: 10.1016/j.ydbio.2004.03.031. [DOI] [PubMed] [Google Scholar]
- Ihmels J., Bergmann S., Barkai N., Bergmann S., Barkai N., Barkai N. Defining transcription modules using large-scale gene expression data. Bioinformatics. 2004;20:1993–2003. doi: 10.1093/bioinformatics/bth166. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Iyer V.R., Horak C.E., Scafe C.S., Botstein D., Snyder M., Brown P.O., Horak C.E., Scafe C.S., Botstein D., Snyder M., Brown P.O., Scafe C.S., Botstein D., Snyder M., Brown P.O., Botstein D., Snyder M., Brown P.O., Snyder M., Brown P.O., Brown P.O. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature. 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- Johansson O., Alkema W., Wasserman W.W., Lagergren J., Alkema W., Wasserman W.W., Lagergren J., Wasserman W.W., Lagergren J., Lagergren J. Identification of functional clusters of transcription factor binding motifs in genome sequences: The MSCAN algorithm. Bioinformatics. 2003;19:i169–i176. doi: 10.1093/bioinformatics/btg1021. [DOI] [PubMed] [Google Scholar]
- Kapranov P., Cawley S.E., Drenkow J., Bekiranov S., Strausberg R.L., Fodor S.P.A., Gingeras T.R., Cawley S.E., Drenkow J., Bekiranov S., Strausberg R.L., Fodor S.P.A., Gingeras T.R., Drenkow J., Bekiranov S., Strausberg R.L., Fodor S.P.A., Gingeras T.R., Bekiranov S., Strausberg R.L., Fodor S.P.A., Gingeras T.R., Strausberg R.L., Fodor S.P.A., Gingeras T.R., Fodor S.P.A., Gingeras T.R., Gingeras T.R. Large-scale transcriptional activity in chromosomes 21 and 22. Science. 2002;296:916–919. doi: 10.1126/science.1068597. [DOI] [PubMed] [Google Scholar]
- Karolchik D., Baertsch R., Diekhans M., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Baertsch R., Diekhans M., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Diekhans M., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., Schwartz M., Sugnet C.W., Thomas D.J., Sugnet C.W., Thomas D.J., Thomas D.J., et al. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama S., Tomaru Y., Kasukawa T., Waki K., Nakanishi M., Nakamura M., Nishida H., Yap C.C., Suzuki M., Kawai J., Tomaru Y., Kasukawa T., Waki K., Nakanishi M., Nakamura M., Nishida H., Yap C.C., Suzuki M., Kawai J., Kasukawa T., Waki K., Nakanishi M., Nakamura M., Nishida H., Yap C.C., Suzuki M., Kawai J., Waki K., Nakanishi M., Nakamura M., Nishida H., Yap C.C., Suzuki M., Kawai J., Nakanishi M., Nakamura M., Nishida H., Yap C.C., Suzuki M., Kawai J., Nakamura M., Nishida H., Yap C.C., Suzuki M., Kawai J., Nishida H., Yap C.C., Suzuki M., Kawai J., Yap C.C., Suzuki M., Kawai J., Suzuki M., Kawai J., Kawai J., et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- Kerber B., Fellert S., Hoch M., Fellert S., Hoch M., Hoch M. Seven-up, the Drosophila homolog of the COUP-TF orphan receptors, controls cell proliferation in the insect kidney. Genes & Dev. 1998;12:1781–1786. doi: 10.1101/gad.12.12.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King D.C., Taylor J., Elnitski L., Chiaromonte F., Miller W., Hardison R.C., Taylor J., Elnitski L., Chiaromonte F., Miller W., Hardison R.C., Elnitski L., Chiaromonte F., Miller W., Hardison R.C., Chiaromonte F., Miller W., Hardison R.C., Miller W., Hardison R.C., Hardison R.C. Evaluation of regulatory potential and conservation scores for detecting cis-regulatory modules in aligned mammalian genome sequences. Genome Res. 2005;15:1051–1060. doi: 10.1101/gr.3642605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloster M., Tang C., Wingreen N.S., Tang C., Wingreen N.S., Wingreen N.S. Finding regulatory modules through large-scale gene-expression data analysis. Bioinformatics. 2005;21:1172–1179. doi: 10.1093/bioinformatics/bti096. [DOI] [PubMed] [Google Scholar]
- Kolbe D., Taylor J., Elnitski L., Eswara P., Li J., Miller W., Hardison R.C., Chiaromonte F., Taylor J., Elnitski L., Eswara P., Li J., Miller W., Hardison R.C., Chiaromonte F., Elnitski L., Eswara P., Li J., Miller W., Hardison R.C., Chiaromonte F., Eswara P., Li J., Miller W., Hardison R.C., Chiaromonte F., Li J., Miller W., Hardison R.C., Chiaromonte F., Miller W., Hardison R.C., Chiaromonte F., Hardison R.C., Chiaromonte F., Chiaromonte F. Regulatory potential scores from genome-wide three-way alignments of human, mouse, and rat. Genome Res. 2004;14:700–707. doi: 10.1101/gr.1976004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolchanov N.A., Ignatieva E.V., Ananko E.A., Podkolodnaya O.A., Stepanenko I.L., Merkulova T.I., Pozdnyakov M.A., Podkolodny N.L., Naumochkin A.N., Romashchenko A.G., Ignatieva E.V., Ananko E.A., Podkolodnaya O.A., Stepanenko I.L., Merkulova T.I., Pozdnyakov M.A., Podkolodny N.L., Naumochkin A.N., Romashchenko A.G., Ananko E.A., Podkolodnaya O.A., Stepanenko I.L., Merkulova T.I., Pozdnyakov M.A., Podkolodny N.L., Naumochkin A.N., Romashchenko A.G., Podkolodnaya O.A., Stepanenko I.L., Merkulova T.I., Pozdnyakov M.A., Podkolodny N.L., Naumochkin A.N., Romashchenko A.G., Stepanenko I.L., Merkulova T.I., Pozdnyakov M.A., Podkolodny N.L., Naumochkin A.N., Romashchenko A.G., Merkulova T.I., Pozdnyakov M.A., Podkolodny N.L., Naumochkin A.N., Romashchenko A.G., Pozdnyakov M.A., Podkolodny N.L., Naumochkin A.N., Romashchenko A.G., Podkolodny N.L., Naumochkin A.N., Romashchenko A.G., Naumochkin A.N., Romashchenko A.G., Romashchenko A.G. Transcription Regulatory Regions Database TRRD: Its status in 2002. Nucleic Acids Res. 2002;30:312–317. doi: 10.1093/nar/30.1.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivan W., Wasserman W.W., Wasserman W.W. A predictive model for regulatory sequences directing liver-specific transcription. Genome Res. 2001;11:1559–1566. doi: 10.1101/gr.180601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laganière J., Deblois G., Lefebvre C., Bataille A.R., Robert F., Giguère V., Deblois G., Lefebvre C., Bataille A.R., Robert F., Giguère V., Lefebvre C., Bataille A.R., Robert F., Giguère V., Bataille A.R., Robert F., Giguère V., Robert F., Giguère V., Giguère V. From the Cover: Location analysis of estrogen receptor α target promoters reveals that FOXA1 defines a domain of the estrogen response. Proc. Natl. Acad. Sci. 2005;102:11651–11656. doi: 10.1073/pnas.0505575102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lantinga-van Leeuwen I.S., Leonhard W.N., Dauwerse H., Baelde H.J., Oost B.A.V., Breuning M.H., Peters D.J.M., Leonhard W.N., Dauwerse H., Baelde H.J., Oost B.A.V., Breuning M.H., Peters D.J.M., Dauwerse H., Baelde H.J., Oost B.A.V., Breuning M.H., Peters D.J.M., Baelde H.J., Oost B.A.V., Breuning M.H., Peters D.J.M., Oost B.A.V., Breuning M.H., Peters D.J.M., Breuning M.H., Peters D.J.M., Peters D.J.M. Common regulatory elements in the polycystic kidney disease 1 and 2 promoter regions. Eur. J. Hum. Genet. 2005;13:649–659. doi: 10.1038/sj.ejhg.5201392. [DOI] [PubMed] [Google Scholar]
- Lee T.I., Rinaldi N.J., Robert F., Odom D.T., Bar-Joseph Z., Gerber G.K., Hannett N.M., Harbison C.T., Thompson C.M., Simon I., Rinaldi N.J., Robert F., Odom D.T., Bar-Joseph Z., Gerber G.K., Hannett N.M., Harbison C.T., Thompson C.M., Simon I., Robert F., Odom D.T., Bar-Joseph Z., Gerber G.K., Hannett N.M., Harbison C.T., Thompson C.M., Simon I., Odom D.T., Bar-Joseph Z., Gerber G.K., Hannett N.M., Harbison C.T., Thompson C.M., Simon I., Bar-Joseph Z., Gerber G.K., Hannett N.M., Harbison C.T., Thompson C.M., Simon I., Gerber G.K., Hannett N.M., Harbison C.T., Thompson C.M., Simon I., Hannett N.M., Harbison C.T., Thompson C.M., Simon I., Harbison C.T., Thompson C.M., Simon I., Thompson C.M., Simon I., Simon I., et al. Transcriptional regulatory networks in. Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- Levine M., Tjian R., Tjian R. Transcription regulation and animal diversity. Nature. 2003;424:147–151. doi: 10.1038/nature01763. [DOI] [PubMed] [Google Scholar]
- Lexa M., Horak J., Brzobohaty B., Horak J., Brzobohaty B., Brzobohaty B. Virtual PCR. Bioinformatics. 2001;17:192–193. doi: 10.1093/bioinformatics/17.2.192. [DOI] [PubMed] [Google Scholar]
- Mach B., Steimle V., Martinez-Soria E., Reith W., Steimle V., Martinez-Soria E., Reith W., Martinez-Soria E., Reith W., Reith W. Regulation of MHC class II genes: Lessons from a disease. Annu. Rev. Immunol. 1996;14:301–331. doi: 10.1146/annurev.immunol.14.1.301. [DOI] [PubMed] [Google Scholar]
- Mantovani R. The molecular biology of the CCAAT-binding factor NF-Y. Gene. 1999;239:15–27. doi: 10.1016/s0378-1119(99)00368-6. [DOI] [PubMed] [Google Scholar]
- Martens J.A., Laprade L., Winston F., Laprade L., Winston F., Winston F. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature. 2004;429:571–574. doi: 10.1038/nature02538. [DOI] [PubMed] [Google Scholar]
- Matys V., Fricke E., Geffers R., Gössling E., Haubrock M., Hehl R., Hornischer K., Karas D., Kel A.E., Kel-Margoulis O.V., Fricke E., Geffers R., Gössling E., Haubrock M., Hehl R., Hornischer K., Karas D., Kel A.E., Kel-Margoulis O.V., Geffers R., Gössling E., Haubrock M., Hehl R., Hornischer K., Karas D., Kel A.E., Kel-Margoulis O.V., Gössling E., Haubrock M., Hehl R., Hornischer K., Karas D., Kel A.E., Kel-Margoulis O.V., Haubrock M., Hehl R., Hornischer K., Karas D., Kel A.E., Kel-Margoulis O.V., Hehl R., Hornischer K., Karas D., Kel A.E., Kel-Margoulis O.V., Hornischer K., Karas D., Kel A.E., Kel-Margoulis O.V., Karas D., Kel A.E., Kel-Margoulis O.V., Kel A.E., Kel-Margoulis O.V., Kel-Margoulis O.V., et al. TRANSFAC: Transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina J.M., Tabernero A., Tabernero A. Astrocyte-synthesized oleic acid behaves as a neurotrophic factor for neurons. J. Physiol. (Paris) 2002;96:265–271. doi: 10.1016/s0928-4257(02)00015-3. [DOI] [PubMed] [Google Scholar]
- Noble W.S., Kuehn S., Thurman R., Yu M., Stamatoyannopoulos J., Kuehn S., Thurman R., Yu M., Stamatoyannopoulos J., Thurman R., Yu M., Stamatoyannopoulos J., Yu M., Stamatoyannopoulos J., Stamatoyannopoulos J. Predicting the in vivo signature of human gene regulatory sequences. Bioinformatics. 2005;21:i338–i343. doi: 10.1093/bioinformatics/bti1047. [DOI] [PubMed] [Google Scholar]
- Petz L.N., Ziegler Y.S., Schultz J.R., Nardulli A.M., Ziegler Y.S., Schultz J.R., Nardulli A.M., Schultz J.R., Nardulli A.M., Nardulli A.M. Fos and Jun inhibit estrogen-induced transcription of the human progesterone receptor gene through an activator protein-1 site. Mol. Endocrinol. 2004;18:521–532. doi: 10.1210/me.2003-0105. [DOI] [PubMed] [Google Scholar]
- Philippakis A.A., He F.S., Bulyk M.L., He F.S., Bulyk M.L., Bulyk M.L.2005Modulefinder: A tool for computational discovery of cis regulatory modules. In Proc. Pac. Symp. Biocomput. 519–530. [PMC free article] [PubMed] [Google Scholar]
- Pravettoni A., Colciago A., Negri-Cesi P., Villa S., Celotti F., Colciago A., Negri-Cesi P., Villa S., Celotti F., Negri-Cesi P., Villa S., Celotti F., Villa S., Celotti F., Celotti F. Ontogenetic development, sexual differentiation, and effects of Aroclor 1254 exposure on expression of the arylhydrocarbon receptor and of the arylhydrocarbon receptor nuclear translocator in the rat hypothalamus. Reprod. Toxicol. 2005;20:521–530. doi: 10.1016/j.reprotox.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Ren B., Robert F., Wyrick J.J., Aparicio O., Jennings E.G., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Robert F., Wyrick J.J., Aparicio O., Jennings E.G., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Wyrick J.J., Aparicio O., Jennings E.G., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Aparicio O., Jennings E.G., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Jennings E.G., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Schreiber J., Hannett N., Kanin E., Hannett N., Kanin E., Kanin E., et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- Robertson G., Bilenky M., Lin K., He A., Yuen W., Dagpinar M., Varhol R., Teague K., Griffith O.L., Zhang X., Bilenky M., Lin K., He A., Yuen W., Dagpinar M., Varhol R., Teague K., Griffith O.L., Zhang X., Lin K., He A., Yuen W., Dagpinar M., Varhol R., Teague K., Griffith O.L., Zhang X., He A., Yuen W., Dagpinar M., Varhol R., Teague K., Griffith O.L., Zhang X., Yuen W., Dagpinar M., Varhol R., Teague K., Griffith O.L., Zhang X., Dagpinar M., Varhol R., Teague K., Griffith O.L., Zhang X., Varhol R., Teague K., Griffith O.L., Zhang X., Teague K., Griffith O.L., Zhang X., Griffith O.L., Zhang X., Zhang X., et al. cisRED: a database system for genome-scale computational discovery of regulatory elements. Nucleic Acids Res. 2006;1:D68–D73. doi: 10.1093/nar/gkj075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S., Skaletsky H., Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Sabo P.J., Humbert R., Hawrylycz M., Wallace J.C., Dorschner M.O., McArthur M., Stamatoyannopoulos J.A., Humbert R., Hawrylycz M., Wallace J.C., Dorschner M.O., McArthur M., Stamatoyannopoulos J.A., Hawrylycz M., Wallace J.C., Dorschner M.O., McArthur M., Stamatoyannopoulos J.A., Wallace J.C., Dorschner M.O., McArthur M., Stamatoyannopoulos J.A., Dorschner M.O., McArthur M., Stamatoyannopoulos J.A., McArthur M., Stamatoyannopoulos J.A., Stamatoyannopoulos J.A. Genome-wide identification of DNaseI hypersensitive sites using active chromatin sequence libraries. Proc. Natl. Acad. Sci. 2004;10:4537–4542. doi: 10.1073/pnas.0400678101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagai T., Hosoya M., Mizushina Y., Tamura M., Shiroishi T., Hosoya M., Mizushina Y., Tamura M., Shiroishi T., Mizushina Y., Tamura M., Shiroishi T., Tamura M., Shiroishi T., Shiroishi T. Elimination of a long-range cis-regulatory module causes complete loss of limb-specific Shh expression and truncation of the mouse limb. Development. 2005;132:797–803. doi: 10.1242/dev.01613. [DOI] [PubMed] [Google Scholar]
- Sandelin A., Alkema W., Engstrom P., Wasserman W.W., Lenhard B., Alkema W., Engstrom P., Wasserman W.W., Lenhard B., Engstrom P., Wasserman W.W., Lenhard B., Wasserman W.W., Lenhard B., Lenhard B. JASPAR: An open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32:D91–D94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E., Sharan R., Sharan R. A discriminative model for identifying spatial cis-regulatory modules. J. Comput. Biol. 2005;12:822–834. doi: 10.1089/cmb.2005.12.822. [DOI] [PubMed] [Google Scholar]
- Segal E., Yelensky R., Koller D., Yelensky R., Koller D., Koller D. Genome-wide discovery of transcriptional modules from DNA sequence and gene expression. Bioinformatics. 2003;19:i273–i282. doi: 10.1093/bioinformatics/btg1038. [DOI] [PubMed] [Google Scholar]
- Sharan R., Ben-Hur A., Loots G.G., Ovcharenko I., Ben-Hur A., Loots G.G., Ovcharenko I., Loots G.G., Ovcharenko I., Ovcharenko I. CREME: Cis regulatory module explorer for the human genome. Nucleic Acids Res. 2004;32:W253–W256. doi: 10.1093/nar/gkh385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrocks A.D. The ETS-domain transcription factor family. Nat. Rev. Mol. Cell Biol. 2001;2:827–837. doi: 10.1038/35099076. [DOI] [PubMed] [Google Scholar]
- Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richard S., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richard S., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richard S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richard S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richard S., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richard S., Clawson H., Spieth J., Hillier L.W., Richard S., Spieth J., Hillier L.W., Richard S., Hillier L.W., Richard S., Richard S., et al. Evolutionarily conserved elements in vertebrates, insects, worms, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S., Nimwegen E.V., Siggia E.D., Nimwegen E.V., Siggia E.D., Siggia E.D. A probabilistic method to detect regulatory modules. Bioinformatics. 2003;19:i292–i301. doi: 10.1093/bioinformatics/btg1040. [DOI] [PubMed] [Google Scholar]
- Sinha S., Schroeder M.D., Unnerstall U., Gaul U., Siggia E.D., Schroeder M.D., Unnerstall U., Gaul U., Siggia E.D., Unnerstall U., Gaul U., Siggia E.D., Gaul U., Siggia E.D., Siggia E.D. Cross-species comparison significantly improves genome-wide prediction of cis-regulatory modules in Drosophila. BMC Bioinformatics. 2004;5:129. doi: 10.1186/1471-2105-5-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek R., Ast G., Ast G. Intronic sequences flanking alternatively spliced exons are conserved between human and mouse. Genome Res. 2003;13:1631–1637. doi: 10.1101/gr.1208803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su A.I., Wiltshire T., Batalov S., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Wiltshire T., Batalov S., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Batalov S., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Zhang J., Soden R., Hayakawa M., Kreiman G., Soden R., Hayakawa M., Kreiman G., Hayakawa M., Kreiman G., Kreiman G., et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapscott S.J. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development. 2005;132:2685–2695. doi: 10.1242/dev.01874. [DOI] [PubMed] [Google Scholar]
- Thomas J.W., Touchman J.W., Blakesley R.W., Bouffard G.G., Beckstrom-Sternberg S.M., Margulies E.H., Blanchette M., Siepel A.C., Thomas P.J., McDowell J.C., Touchman J.W., Blakesley R.W., Bouffard G.G., Beckstrom-Sternberg S.M., Margulies E.H., Blanchette M., Siepel A.C., Thomas P.J., McDowell J.C., Blakesley R.W., Bouffard G.G., Beckstrom-Sternberg S.M., Margulies E.H., Blanchette M., Siepel A.C., Thomas P.J., McDowell J.C., Bouffard G.G., Beckstrom-Sternberg S.M., Margulies E.H., Blanchette M., Siepel A.C., Thomas P.J., McDowell J.C., Beckstrom-Sternberg S.M., Margulies E.H., Blanchette M., Siepel A.C., Thomas P.J., McDowell J.C., Margulies E.H., Blanchette M., Siepel A.C., Thomas P.J., McDowell J.C., Blanchette M., Siepel A.C., Thomas P.J., McDowell J.C., Siepel A.C., Thomas P.J., McDowell J.C., Thomas P.J., McDowell J.C., McDowell J.C., et al. Comparative analyses of multi-species sequences from targeted genomic regions. Nature. 2003;424:788–793. doi: 10.1038/nature01858. [DOI] [PubMed] [Google Scholar]
- Thompson W., Palumbo M.J., Wasserman W.W., Liu J.S., Lawrence C.E., Palumbo M.J., Wasserman W.W., Liu J.S., Lawrence C.E., Wasserman W.W., Liu J.S., Lawrence C.E., Liu J.S., Lawrence C.E., Lawrence C.E. Decoding human regulatory circuits. Genome Res. 2004;14:1967–1974. doi: 10.1101/gr.2589004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viatour P., Merville M., Bours V., Chariot A., Merville M., Bours V., Chariot A., Bours V., Chariot A., Chariot A. Phosphorylation of NF-κB and IκB proteins: Implications in cancer and inflammation. Trends Biochem. Sci. 2005;30:43–52. doi: 10.1016/j.tibs.2004.11.009. [DOI] [PubMed] [Google Scholar]
- Wang W., Cherry J.M., Nochomovitz Y., Jolly E., Botstein D., Li H., Cherry J.M., Nochomovitz Y., Jolly E., Botstein D., Li H., Nochomovitz Y., Jolly E., Botstein D., Li H., Jolly E., Botstein D., Li H., Botstein D., Li H., Li H. Inference of combinatorial regulation in yeast transcriptional networks: A case study of sporulation. Proc. Natl. Acad. Sci. 2005;102:1998–2003. doi: 10.1073/pnas.0405537102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman W.W., Fickett J.W., Fickett J.W. Identification of regulatory regions which confer muscle-specific gene expression. J. Mol. Biol. 1998;278:167–181. doi: 10.1006/jmbi.1998.1700. [DOI] [PubMed] [Google Scholar]
- Woolfe A., Goodson M., Goode D.K., Snell P., McEwen G.K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K., Goodson M., Goode D.K., Snell P., McEwen G.K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K., Goode D.K., Snell P., McEwen G.K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K., Snell P., McEwen G.K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K., McEwen G.K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K., Smith S.F., North P., Callaway H., Kelly K., North P., Callaway H., Kelly K., Callaway H., Kelly K., Kelly K., et al. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlinger J., Simon I., Harbison C.T., Hannett N.M., Volkert T.L., Fink G.R., Young R.A., Simon I., Harbison C.T., Hannett N.M., Volkert T.L., Fink G.R., Young R.A., Harbison C.T., Hannett N.M., Volkert T.L., Fink G.R., Young R.A., Hannett N.M., Volkert T.L., Fink G.R., Young R.A., Volkert T.L., Fink G.R., Young R.A., Fink G.R., Young R.A., Young R.A. Program-specific distribution of a transcription factor dependent on partner transcription factor and MAPK signaling. Cell. 2003;113:395–404. doi: 10.1016/s0092-8674(03)00301-5. [DOI] [PubMed] [Google Scholar]
- Zhou Q., Wong W.H., Wong W.H. CisModule: De novo discovery of cis-regulatory modules by hierarchical mixture modeling. Proc. Natl. Acad. Sci. 2004;101:12114–12119. doi: 10.1073/pnas.0402858101. [DOI] [PMC free article] [PubMed] [Google Scholar]