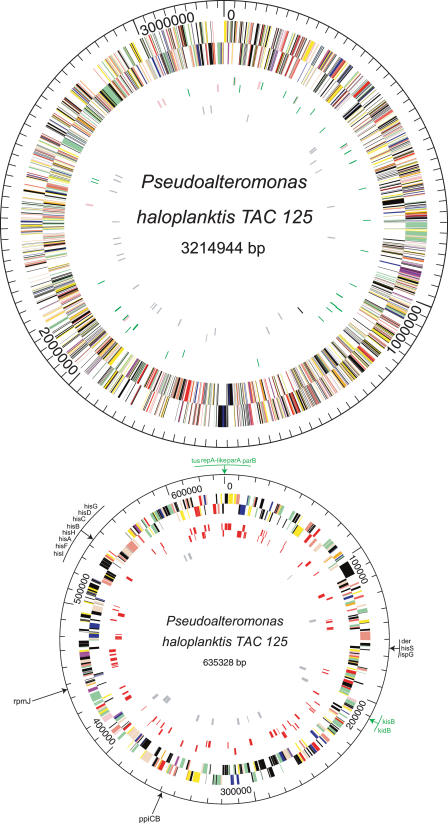
Figure 1.
Circular representation of the Pseudoalteromonas haloplanktis genome. Circles display (from the outside): (1) predicted coding regions transcribed in the clockwise direction; (2) predicted coding regions transcribed in the counterclockwise direction. Genes displayed in 1 and 2 are color-coded according to different functional categories: salmon indicates amino acid biosynthesis; orange indicates purines, pyrimidines, nucleosides, and nucleotides; purple indicates fatty acid and phospholipid metabolism; light blue indicates biosynthesis of cofactors, prosthetic groups, and carriers; light green indicates cell envelope; red indicates cellular processes; brown indicates central intermediary metabolism; yellow indicates DNA metabolism; green indicates energy metabolism; pink indicates protein fate/synthesis; blue indicates regulatory functions; grey indicates transcription; teal indicates transport and binding proteins; and black indicates hypothetical and conserved hypothetical proteins. (3) tRNAs (green) and rRNA (pink) on chrI/genes similar to phage proteins (red) on chrII; (4) and tonB and tonB-like genes in grey. Chromosome II gene names similar to that of the R1 plasmid replication apparatus (unidirectional) are colored in green.