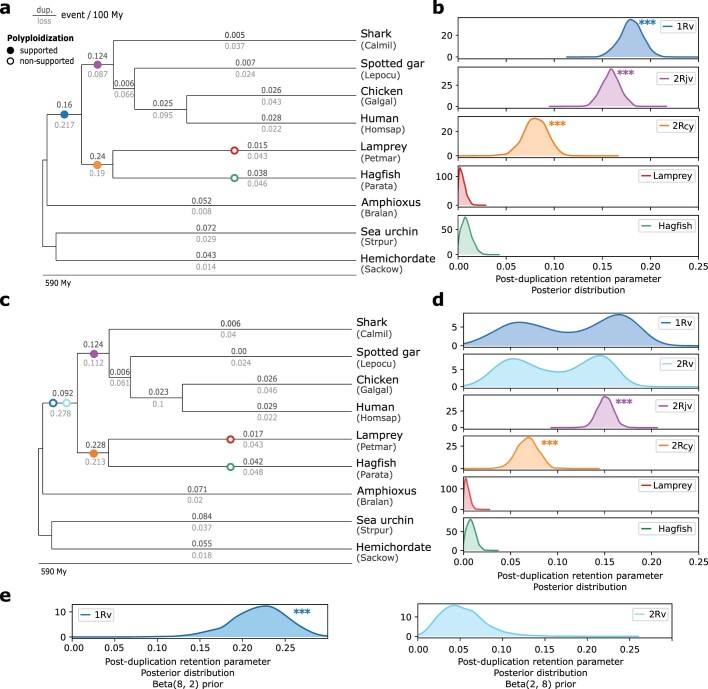
Extended Data Fig. 4. Tests of genome duplication hypotheses on the vertebrate tree.
a, Species phylogeny and polyploidization hypotheses tested with WHALE36 using 8,931 gene families (Methods, see Supplementary Table 8 for details of the genomes used in the analysis). Polyploidization hypotheses are indicated by circles on the corresponding branches, with supported polyploidizations indicated with solid circles. Inferred background gene duplication and loss rates are presented on the branches. b, Posterior distribution obtained for the WHALE post-duplication retention parameter q, for each hypothesis presented in a. Stars indicate distributions significantly different from 0 (Bayes factors BFNull_vs_WGD < 10−3), which correspond to the supported polyploidization events. c, Alternative set of polyploidization hypotheses tested, as in a, but with two successive duplications proposed in the ancestral vertebrate lineage (1RV and 2RV). d, Posterior distribution obtained for the WHALE post-duplication retention parameter, for each hypothesis presented in c. Here, the posterior distribution for retention parameters of the 1RV and 2RV events are bimodal, suggesting that the method cannot effectively separate parameters estimated for 1RV and 2RV when starting from identical priors. e, Use of distinct priors on 1RV (Beta(8, 2)) and 2RV (Beta(2, 8)) separates the estimated posterior distribution into distinct unimodal posterior distributions and provides support for a single shared 1Rv event in the vertebrate stem lineage. This analysis was performed on a random subset of 1,000 gene families, to reduce computational time (Methods).