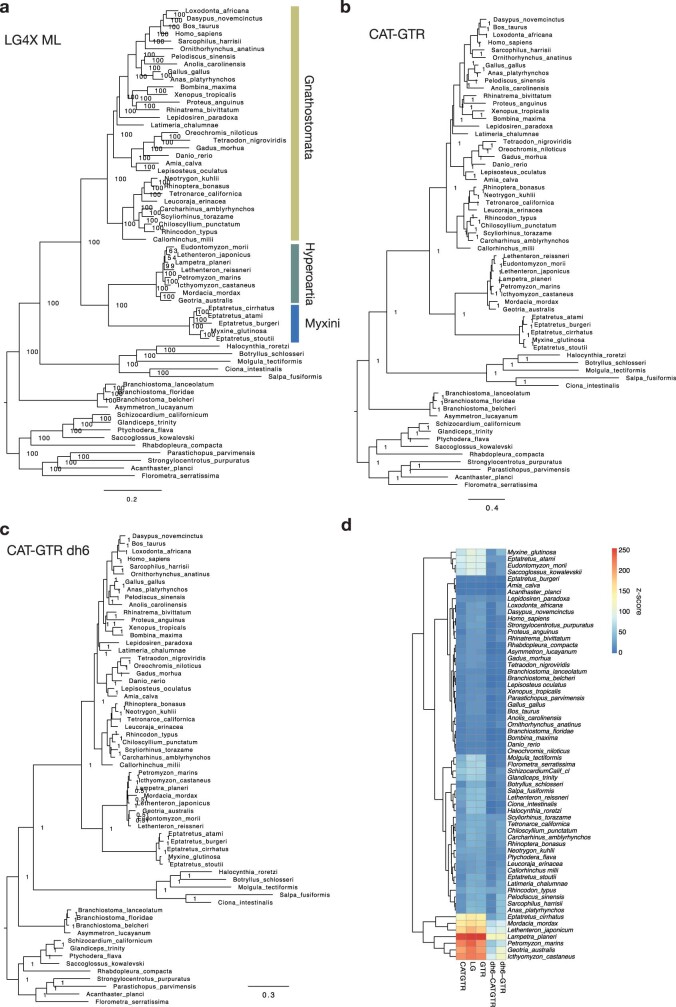
Extended Data Fig. 2. Phylogenetic reconstruction of deuterostome relationships with a focus on cyclostome position.
a, Tree reconstructed with IQ-TREE assuming LG4X model using a dataset of 1,467 single-copy orthologues and a partitioned model. b, Tree reconstructed using PhyloBayes and a CAT+GTR+G4 model using a subset of 176 orthologues showing the lowest saturation (see methods). c, Tree reconstructed using the same set of orthologues after Dayhoff 6 categories amino acid recoding to account for possible compositional heterogeneity due to high GC% in cyclostome genomes. d, z-score of posterior predictive analyses to assess composition heterogeneity. Positive z-scores indicate that average amino acid diversity is underestimated (negative z-scores indicate an overestimation) which highlights the composition bias existing in some lamprey and hagfish species and shows that recoding (Dayhoff 6) alleviates these biases.