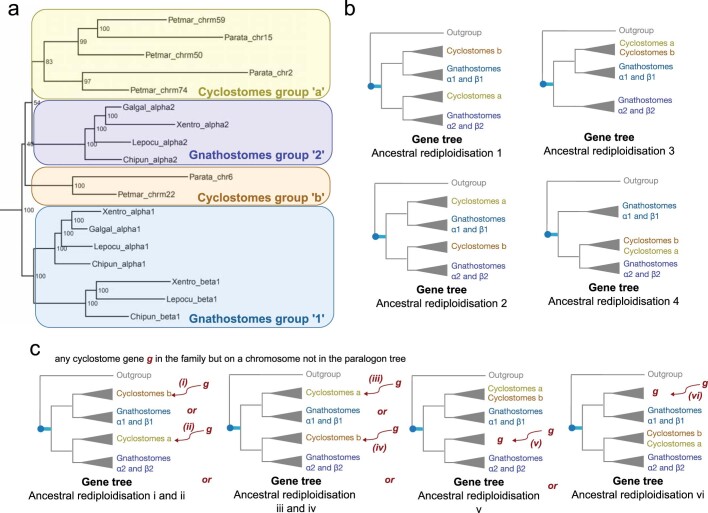
Extended Data Fig. 6. Method for construction of post-1R ancestral rediploidization constrained gene-tree topologies, using CLGM as an example.
a, Gnathostome 1R 1 and 1R 2 copies can be confidently identified and serve as a skeleton to build ancestral rediploidization tree topologies (blue-purple groups). Hagfish and lamprey chromosomes confidently grouped in a clade from the CLGM paralogon tree are defined as potential 1R-derived paralogons (yellow-orange groups) and kept together in the constrained ancestral rediploidization tree topology (see b). All sets of cyclostome chromosomes that were kept together for other CLGs are indicated in Supplementary Table 5. b, Possible groupings of hagfish and lamprey genes with gnathostome genes based on their chromosomal location, following 1R ancestral rediploidization. c. Genes located on hagfish and lamprey chromosomes that are not considered in the reconstructed paralogon tree (due to low representation because of small-scale rearrangements displacing them on different chromosomes) can each be placed on either side of the duplication in the absence of any prior information. In the presented scenario, this results in six different possible ancestral rediploidization (i to vi) constrained tree topologies. Only topologies with a maximum of three lamprey genes and three hagfish genes on each side of the 1R are permitted, to remove possibly confounding effects of complex multicopy gene families.